Abstract
The diagnostic classification of digitized tissue images based on histopathologic lesions present in whole slide images (WSI) is a significant task that eludes modern image classification techniques. Even with advanced methods designed for digital histopathology, the domain of toxicologic pathology presents challenges in that histopathologic features may be at times complex, subtle, and/or rare. We propose an innovative weakly supervised learning method that leverages minimal annotations, a state-of-the-art self-supervised vision transformer for embedding extraction, and a novel guided attention mechanism that is better suited for heavily imbalanced datasets typical in toxicologic pathology. Our model demonstrates improvements in diagnostic classification and attention heatmap quality over the previously described clustering-constrained-attention multiple-instance learning method on several lesion classes in rat livers (38% improvement in AUC). We also demonstrate how an ensemble of binary classifiers improves interpretability and allows for multiclass classification and the classification of diagnostic regions of interest in each slide. The improved classification performance and higher contrast heatmaps better support toxicologic pathologists’ histopathology analysis and will enable more efficient workflows as they are further refined and integrated into routine use.
Similar content being viewed by others
Introduction
Identifying lesions in whole slide images (WSIs) of organ tissues is a critical objective in toxicologic histopathology (TOXPATH). Typically, this screening process is achieved manually by experienced pathologists who examine each glass tissue slide and render a diagnosis based on each observed tissue characteristic. This process, while time consuming, remains the gold standard analysis method. The transformation of pathology into a digital medium by the development and widespread use of whole WSI digital scanning methods allows for computational advancements to improve the efficiency, consistency, and robustness of analysis. Particularly, developments in the field of artificial intelligence have allowed methods such as deep learning techniques to be applied towards tasks such as image classification1,2, semantic segmentation3,4,5, outcome prediction6,7and anomaly detection8,9.
However, the field of TOXPATH presents several challenges that complicate the direct translation of such deep learning techniques to digital histopathology images. One such technical issue is large image sizes, with a single WSI consisting of billions of pixels and gigabytes of data. The memory limitations of current devices preclude analysis of such large files and necessitate alternative solutions such as tiling whole slide images into smaller, processable patches8,10,11. Another difficulty is procuring large numbers of high-quality pixel-level contour annotations of WSIs that highlight features of interest such as pathologic lesions. Often, only limited semantic information is available such as a single label to characterize the entire WSI, which demands methods such as weak-supervision rather than fully supervised learning algorithms8,11,12,13,14. Despite the large quantities of data when measured in terms of pixels and bytes, the challenge in the domain of TOXPATH is the limited number of available WSIs and corresponding diagnostic labels, particularly representing rare lesion classes. A single dataset consisting of terabytes of information may only be composed of approximately a thousand WSIs and their corresponding diagnostic labels; this data limitation is a barrier in the development of robust algorithms for tissue-level classification15,16.
Additional challenges characterize histopathological data, especially in the context of domains such as TOXPATH. The underlying data is highly complex; tissue comprises multiple diverse structural and functional constituents such as organ functional parenchyma, multiple types and sizes of vessels, various extracellular matrix components, and other supporting and functional elements, all arranged in highly structured and heterogeneous patterns in three dimensions. Any single histology section of tissue, typically 3–5 microns in thickness, is essentially a two-dimensional sample, taken non-uniformly from this three-dimensional tissue, and will contain tissue structures sampled from different regions in a wide variety of planes, resulting in highly incongruous samples. Specifically, in the WSIs used in this study, the histopathologic features that distinguish class instances are quite subtle, with many mild or minimal instances as opposed to more obvious, severe lesions. Tissues affected by a lesion will typically have a relatively small percentage of tissue affected, which leads to a heavy class imbalance where the vast majority of tissue is normal. This imbalance is further exacerbated by the fact that most tissues are lesion-free. Despite this imbalance, it remains critical to detect and classify the abnormal incidences that can exhibit themselves sparsely within a tissue and with many different representations among tissue sections. In sum, TOXPATH data requires innovative computational approaches to address the complexity of the samples and to contend with the heavy class imbalance inherent in TOXPATH8,11,12,17.
Some recent applications of deep learning to pathology data have shown promising results on WSIs that have an accompanying label that denotes diagnostic characteristic classes11,14,17,18. Methods that leverage multiple-instance-learning can take advantage of the WSI labels as weak supervision to achieve respectable WSI classification performance on tasks such as tumor classification and localization11,17. However, we show that there is still room for improvement for domains with heavily healthy/normal-skewed data such as TOXPATH.
Multiple instance learning (MIL) relies on tile level features that are extracted by a pretrained neural network. Typically, a convolutional feature extractor pretrained on a dataset like ImageNet is used to reduce each tile into a lower-dimensional embedding. However, as ImageNet contains ordinary images, it is unclear if the features learned by the feature extractor generalize to histology images. Self-supervised learning methods offer a solution for the data density and label sparsity challenges inherent to pathology datasets. In self-supervised learning, a pretext task is used to learn a feature extractor in a completely label-free manner19. This feature extractor can be used for downstream tasks including multiple instance learning-based image classification. Self-supervised learning has been used for feature extraction in digital pathology for tasks including semantic segmentation and image classification. In dual stream multiple instance learning, a self-supervised convolutional network was used for feature extraction20,21. Other approaches have successfully used self-supervised learning to train feature extractors tailored to histopathology data22,23. Recently, self-supervised learning has been shown to be highly effective for training large vision transformer models, which have been shown to outperform convolutional networks on many tasks20,24,25 and are beginning to see use in digital pathology26,27,28,29. However, these techniques have not yet been applied to TOXPATH and we sought to evaluate different feature extractors in our models for TOXPATH datasets.
Multiple instance learning methods typically aggregate tile level features into whole slide level features. The model then learns to predict WSI labels based on the aggregated features11. Choice of aggregation mechanism is a crucial design choice for MIL models. The simplest aggregation function is to take the mean or maximum of tile level features. In attention MIL15, the aggregation function is a weighted average of the tile level features where the weights are learned by a neural network, while in TransMIL17 self-attention is used to learn spatial relationships between embedded patches and aggregate a single whole slide embedding. Other methods use more complicated aggregations to explicitly model sparse relationships between patches30. Attention scores can be visualized as heatmaps to highlight high attention areas of diagnostic interest. Clustering-constrained-attention multiple-instance learning (CLAM)11 improves on attention MIL by learning attention scores and requiring that the highest and lowest scored features be linearly separable. In doing so, CLAM encourages contrast between normal and abnormal tissue scores which results in high contrast heatmaps. CLAM simultaneously aggregates all patch-level representations into a WSI-level representation, which allows all patches in a WSI to contribute to the final class prediction. In contrast, traditional multiple instance learning methods are restricted in that they use alternative aggregation functions such as max-pooling to determine a single instance or patch to best represent the entire WSI1. However, this method relies on the assumption that both normal and abnormal tissue will be present in a given tissue image, which is often not the case in the predominantly normal tissue common to TOXPATH datasets. We therefore desire an attention mechanism that increases contrast in abnormal WSIs and reduces contrast in normal WSIs. Improvements to current methods are required for application to TOXPATH datasets.
Our method extends current attention-based multiple-instance-learning methods of weakly supervised WSI classification11. We leverage a self-supervised vision transformer24 to obtain comprehensive embeddings of patches within a WSI that allow for the representation of the WSI in a lower dimension space. We then apply an attention-mechanism to the embeddings to differentiate relevant and non-relevant patches. The attention score and the embeddings are used in the downstream task of classification that is weakly supervised by a slide-level label.
To specifically address the normal/lesioned tissue data imbalance, an attention-mining loss is used to self-supervise and guide the attention of the network to the most relevant tiles to improve both classification performance and visualization quality. Our method is notable in that we address imbalance via a novel optimization algorithm, while other approaches in literature use approaches such as data augmentation31 or tailored data sampling strategies32,33.
We utilize a TOXPATH dataset of rat livers containing diverse lesions, reporting binary whole slide classification performance in each of 5 lesions classes and overall lesion detection performance across all classes, to gauge our model’s utility in typical lesion detection workflows. This dataset contains lesions that are typical in pathology workflows and exhibit more severe data imbalance than previous studies10,32,34. We demonstrate that an ensemble of models for each lesion can provide more comprehensive support to pathologists rather than strict binary classification.
Results
We present results for 5 model variations. CLAM is the baseline with a Resnet50 extractor. Models with “G” in the acronym denote models with our guidance mechanism. Models with “CD” denote “custom dino”, a vision transformer trained on in house data using DINO. Finally, DAM and DGAM both use vision transformers trained with DINO on ImageNet.
Improvement of binary diagnostic classification using custom embeddings across multiple datasets
The performance of each model on each dataset measured by AUC is summarized in Table 1. We see that in all datasets except the extremely high variance necrosis dataset, the DINO trained vision transformer offers performance improvements in all cases compared to the Resnet50 model used in CLAM. To enable a thorough comparison between embedding methods, Table 1 presents the results of five different encoders trained under various datasets and training algorithms. We see that self supervision improves performance compared to full supervision on ImageNet for Resnet50. Additionally, we see that the improvement from CLAM to DAM cannot only be attributed to the increased complexity of the ViT encoder compared to Resnet50. Indeed, when the Resnet50 is trained with self supervision, it outperforms ViT with the same pretraining set up (DGAM).
We also observe that our models with feature extractors trained on in house data outperforms CTransPath23, a vision transformer self supervised on TCGA. Figures 1, 2, 3 and 4 show the AUROCs and representative results of normal WSIs and WSIs with diagnosis for all model respectively. Moreover, Table 1 also shows the relative difficulty of each dataset with vacuolation the most accurately classified and ‘all lesions’ being the most poorly classified.
Performance and representative results across different models on the necrosis dataset. The highest performing model is DAM with a mean AUROC of 0.877 ± 0.098. The remaining performances are: 0.820 ± 0.092 (CLAM), 0.824 ± 0.113 (DGAM), 0.842 ± 0.107 (CDAM) and 0.803 ± 0.120 (CGDAM). CDGAM yields the heatmaps with highest segmentation and contrast quality. Note that CLAM, DAM, DGAM and CDAM highlight many tiles from glass and red blood cells whereas our CDGAM highlights liver tissues which contributes to the diagnoses.
Performance and representative results across different models on the vacuolation dataset. The highest performing models are CDAM and CDGAM with a mean AUROC of 0.945 ± 0.021 and 0. 945 ± 0.014 respectively. The remaining performances are: 0.855 ± 0.099 (CLAM), 0.936 ± 0.036 (DAM), and 0.934 ± 0.021 (DGAM). CDGAM yields the heatmaps with highest segmentation and contrast quality. Note that CLAM, DAM, DGAM and CDAM highlight many tiles from glass and red blood cells whereas our CDGAM highlights liver tissues which contributes to the diagnoses.
Performance and representative results across different models on the inflammation dataset. The highest performing model is CDAM with a mean AUROC of 0.866 ± 0.029. The remaining performances are: 0.589 ± 0.035 (CLAM), 0.842 ± 0.036 (DAM), 0.838 ± 0.037 (DGAM), and 0.859 ± 0.027 (CDGAM). CDGAM yields the heatmaps with highest segmentation and contrast quality. Note that CLAM highlights many tiles from glass whereas our CDGAM highlights liver tissues which contributes to the diagnoses.
Performance and representative results across different models on all lesions dataset. The highest performing model is CDGAM with a mean AUROC of 0.815 ± 0.032. The remaining performances are: 0.588 ± 0.047 (CLAM), 0.800 ± 0.038 (DAM), 0.810 ± 0.039 (DGAM), and 0.813 ± 0.041 (CDAM). CDGAM yields the heatmaps with highest segmentation and contrast quality. Note that CLAM highlights many tiles from glass whereas our CDGAM highlights liver tissues which contributes to the diagnoses.
From Table 1, we conclude that self-supervised vision transformers are a universal improvement over conventionally trained Resnet feature extractors. However, we did not detect statistically significant improvement between Imagenet trained ViT and our in house ViT. This implies that the gains we are observing are due to the ViT architecture rather than domain specific pretraining. We also observed that while it does not necessarily improve AUC scores in every case, our guided attention mechanism sharpens heatmap values and better attends to lesioned areas (Fig. 5).
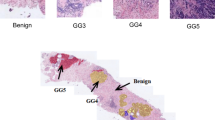
Example heatmaps with annotated pixel level ground truths. We can see that the guidance mechanism (denoted by models with “G” in their name, underlined) leads to qualitatively higher contrast with regards to the ground truth annotation. All proposed models perform better than the CLAM model, which entirely fails to identify the lesions.
Improving classification and visualization of necrosis using custom embeddings and guided attention
The performance of the models on the necrosis dataset are summarized in Fig. 1. DAM has the highest mean test AUROC of 0.877 ± 0.098, which surpasses baseline performance of CLAM of 0.820 ± 0.092. DGAM and CDAM achieve higher results than CLAM at 0.824 ± 0.113 and 0.842 ± 0.107 respectively. CDGAM is the only model that performs more poorly than CLAM at 0.803 ± 0.120. One important consideration is the high variance throughout all models, which we attribute to the highly imbalanced necrosis dataset, with only 115 total necrosis WSIs (12 for testing). We also observe a positive skew in all the ROC curves, which indicates that low false-positive rates are associated with low true-positive rates; the implication is that the models are biased towards more negative, normal predictions, which corresponds to the imbalanced datasets. Despite having relatively few examples of necrosis to train on, Fig. 5 shows that we still achieve high contrast heatmaps, which illustrates the data efficiency of our method. Specifically, our guided attention model (CGDAM) produces the heatmap with the highest contrast, despite the lowest numerical performance. Furthermore, the models that utilize guided attention demonstrate higher contrast than the models without guided attention (Fig. 5). Overall, for the necrosis dataset, we see that the vision transformer provides higher AUC scores compared to the baseline while the guided attention appears to improve heatmap contrast.
Improvement of classification and visualization of vacuolation using custom embeddings and guided attention
Figure 2 summarizes the performance of the model on the vacuolation dataset. CDAM and CDGAM have equally high performance of 0.945 ± 0.021 and 0.945 ± 0.014 respectively, and both exceed the baseline performance of 0.855 ± 0.099. DAM and DGAM also outperform the baseline at 0.936 ± 0.036 and 0.934 ± 0.021 respectively. The vacuolation dataset is more balanced than the necrosis dataset, but still quite imbalanced, which may contribute to the high variance of CLAM performance. We observe a similar pattern as the necrosis results when observing the heatmaps. CLAM erroneously associates non-tissue tiles with high attention. DAM, DGAM, CDAM, and CDGAM all pay high attention to tiles with vacuolation in the abnormal tissue; furthermore, even when they correctly classify the normal tissues, they are still able to suggest tiles with potential vacuolation. Both CDAM and DAM associate non-tissue tiles and blood with low attention, as desired. Several of the heatmaps exhibit gridlines that we believe correspond to artifacts introduced during slide scanning and stitching18,35. This attention towards image stitching artifacts is not ideal as it may distract from the lesions of interest. However, these stitching artifacts are much less pronounced in the guided models, and despite the presence of these artifacts, the overall WSI classification performance in the guided models still outperform CLAM.
Improvement classification and visualization of inflammation using custom embeddings and guided attention
Figure 3 compiles the performance of the models on the inflammation dataset. CDAM has the highest performance of 0.866 ± 0.029 and CDGAM has the second highest performance of 0.859 ± 0.027. DGAM and DAM have performances of 0.838 ± 0.037 and 0.842 ± 0.036 respectively. All models outperform CLAM at 0.589 ± 0.035. The inflammation dataset is far more balanced than the necrosis and vacuolation datasets, which demonstrates that CLAM struggles on more realistically balanced datasets. CLAM associates the non-tissue tiles with high attention. The other models pay attention to tiles with inflammation in the abnormal WSI, correctly identify normal tiles in both normal and abnormal WSIs and suggest abnormal tiles in correctly classified normal tissue sections.
Improved classification and visualization of multiple lesions using custom embeddings and guided attention
The performance of the models on the dataset with all lesions are summarized in Fig. 4. CDGAM has the highest performance of 0.815 ± 0.032. CDAM, DGAM, and DAM achieve performances 0.813 ± 0.04, 0.810 ± 0.039, and 0.800 ± 0.038 respectively. CLAM again performs more poorly at 0.588 ± 0.047. The dataset with all lesions is the most balanced and challenging dataset, since it contains a diverse set of lesions and is a super set of all other datasets combined. This performance enhancement on such a challenging dataset highlights our method’s improvements over CLAM. The normal tissue sections have less signal on the heatmaps than the abnormal tissues, but high attention regions are still highlighted. DGAM and CDAM misclassify the normal tissue, whereas the other models correctly classify the tissue with low confidence. CLAM again associates non-tissue tiles with high attention.
Heatmap contrast improvements using guided attention
We observed qualitatively higher contrast in heatmaps produced by models trained with guided attention. To quantify this improvement in contrast, we annotated 25 ROIs in 5 WSIs presenting necrosis, infiltrate, and vacuolation. We then computed the ratio between mean heatmap pixel intensity in the annotated and unannotated regions. Figure 5 shows examples of annotated regions along with the corresponding heatmaps for each model -- higher agreement is clearly seen when using the proposed attention mechanism. Additionally, the guidance mechanism leads to higher contrast in the annotated regions (see Supplementary Table S1). Using a Wilcoxon non-parametric test, the guidance mechanism leads to statistically significant higher contrast (p << 0.01).
Statistical analysis
To evaluate the impact of encoder choice and guidance mechanism, we perform an ANCOVA with these two variables and test set auc as the response variable. We repeated this analysis in all four datasets. In each case, we failed to detect a statistically significant effect from our guidance mechanism. However, we found that encoder choice had a significant effect, summarized in Supplementary Table S4. We see significant improvements between our in-house trained ViT and the Resnet50 baseline on all datasets except necrosis (p << 0.01). However, we did not detect a significant difference between our in house ViT and ImageNet ViT.
Utility of an ensemble approach
Lastly, Supplementary Fig. S1 demonstrates the utility of an ensemble approach for multi-class classification. After training a CDGAM model on each of the 4 datasets, we apply the models to unseen tissue sections with multiple diagnoses. For the WSIs with necrosis and inflammation, the model correctly classifies the WSIs as abnormal with necrosis and inflammation, without vacuolation. On the other hand, the WSI with only vacuolation is correctly classified as abnormal with vacuolation by the model trained on all lesions and the model trained on vacuolation; but the models trained on necrosis and inflammation correctly do not indicate the presence of either lesion. The final row in Supplementary Fig. S1 a WSI with all three lesions, necrosis, vacuolation, and inflammation, which the models correctly predict.
While the ensemble of binary classifiers performs well on our data, this approach does not scale well when more lesion classes are considered. Indeed, to expand the ensemble approach to more classes, a new model must be trained for each new class. However, feature extraction needs to only be performed once, regardless of the number of classes. To justify our ensemble method, we train a CDGAM model in a multiclass/multilabel manner, taking particular care to ensure that normal predictions are mutually exclusive with lesion classes. As we can see from Supplementary Table S3, the ensemble of models outperforms the multiclass model.
Discussion
We have developed an improved whole slide image tissue classification method based on weakly supervised learning and guided attention mechanisms. Our models outperform the baseline CLAM model in many cases.
The most significant improvements are a result of leveraging vision transformers trained with DINO to reduce patches of whole slide images into lower-dimensional feature embeddings. We observed the most improvement in the more challenging datasets (inflammation and all lesions) where CLAM fails to classify WSIs and produce salient attention heatmaps. Our method better models these complicated and highly variant datasets. Specifically, using a vision transformer trained on histology data yields improvement on all datasets except for the necrosis dataset, which is challenging to assess due the paucity of positive samples resulting in a skewed evaluation metric. Although CLAM outperformed CDGAM in the necrosis dataset, we can still see the improvements afforded by our methods in their enhanced heatmap contrast and lesion attention as compared to CLAM. This underperformance is a direction for future investigation and additional research into attention mining is an area for further development36. From our statistical analysis, we see that most improvement comes from the change to ViT architecture rather than domain specific pretraining.
The use of guided-attention mechanisms significantly improves heatmap quality via contrast enhancement and localized delineation of highly attended regions of interest. However, it has no significant effect on overall slide level classification, measured by test set AUC. In the necrosis dataset, for example, we see that guided attention generates heatmaps with extremely well-delineated abnormal regions and high contrast. Guided attention models (DGAM and CDGAM) are particularly well suited for TOXPATH since many tissue sections would contain only normal tissue without normal-lesion contrast. Of the proposed methods, CDGAM has the highest overall performance. Specifically, we see that CDGAM outperforms all other methods in all lesions dataset, which most resembles the common task of identifying normal tissue from all other classes of lesion. We observed that the guidance mechanism greatly increases contrast between lesioned and non-lesioned tissue and attends more sharply to lesioned areas (Fig. 5). Contrast could be further boosted with conventional nonlinear heatmap normalization techniques and the design and validation of such a transform is a promising direction for future work. Such a choice of nonlinear function is not necessarily straightforward, however, and the fact that our method increases contrast between lesioned and non-lesioned regions without additional calibration is a significant advantage. Using models trained with guided-attention enables greater explainability of algorithm output in that the heatmaps provide human-interpretable insight into the basis for the algorithm results. These heatmaps can additionally help draw pathologists’ attention to suspicious regions of WSIs for expert review. Ensemble models can further support a more comprehensive analysis of whole slide images rather than strict binary classification of a single lesion. While compute/parameters scale linearly with the number of lesion classes (number of models in the ensemble), these classifiers can be run in parallel once tile level feature extraction has been completed. Additionally, feature extraction, which is the most expensive step in our pipeline, is only done once. We consider this increased compute an acceptable tradeoff for the greater degree of interpretability that an ensemble affords.
A significant challenge regarding our in-house data is the intra-dataset variation. A single type of lesion such as necrosis can present with varying histomorphology, and the ground truth diagnoses were made by multiple pathologists in a variety of contexts. A further challenge regarding our data is the varying degrees of severity, ranging from minimal, mild, moderate, and severe. Lesions of different severity can appear drastically different, especially between minimal and severe. Future work can focus on identifying differences in severity and improve performance on the less obvious cases. Another area of improvement is that the heatmaps can be noisy, with some models focusing on regions such as blood or glass that are not relevant to the classification. The datasets can also be refined to improve performance on regions such as blood and glass, which should not receive high attention.
One concern with our work is potential data leakage between our pretraining data and MIL data. Recent work30 has found that this transductive learning configuration could artificially inflate model performance. We found the leakage to be minimal. In total, 14 slides from the pretraining set leaked into the training set (1% of training data). Of these 14 slides, 8 were normal and 6 contained lesions. Additionally, we sample 10,000 tiles randomly per slide during pretraining to assemble our datasets. This sparse sampling further mitigates the effect of a data leak. Given our cross-validation ratio, we can expect 1.4 slides on average to leak into the validation and test sets for each fold. Moreover, models that were not pretrained on our in-house data do not have this data leakage problem and still show strong performance with our guided attention mechanism. Given the limited scope of the data leakage, we do not anticipate this substantially impacting our results. However, investigating the impact of transduction is a valuable direction for future research.
We envision several ways in which our method could greatly improve pathologist slide review workflows. First, pathologists could leverage the method to prioritize the order of their slide review in order to focus their attention on highly important lesion containing WSIs. Second, while reviewing a WSI, pathologists can utilize our heatmaps to guide them quickly to areas with high probability of lesions to identify and focus on these high-impact regions efficiently. We anticipate our method will greatly accelerate pathologist throughput during routine slide review and preserve pathologist bandwidth for the important overall interpretation of study findings. At this time, we do not believe our anomaly detection method can replace pathologists’ review of all WSIs from a safety study, but we envision a future where we may be able to sort normal from abnormal WSIs and greatly reduce the number of tissues a pathologist is required to review.
Future directions include adapting the method to different tissues. The method is not tissue specific, and performance can be evaluated on different organs after curating appropriate datasets. Lastly, we can improve upon the ensemble approach and implement a multi-label model to increase efficiency. We showed that this approach is viable, but still underperforms an ensemble of binary classifiers. More research is required to address this gap.
Methods
Datasets
To create datasets for pretraining, a collection of 120 rat tissue slides collected from across our archive was selected. These slides were generated as part of routine TOXPATH studies between 2008 and 2020. This dataset contains both healthy and lesioned tissue from 5 different organs commonly used in TOXPATH screening. For each slide, we segment the tissue regions from the glass and select 10,000 non overlapping 256pix tiles at 40× from the tissue region. In total, we train on 1.2 m tissue tiles during pretraining.
For training our multiple instance models, the data consists of 2402 in-house rat liver WSIs with WSI-level labels provided by expert pathologists, collected from toxicity studies between 2008 and 2020. We group the 2402 WSIs into different datasets to evaluate our method on different lesions of interest. The three selected lesions of interest are necrosis (n = 115), vacuolation (n = 224), and inflammation (n = 565) that were selected based on the high number of instances within the 2402 WSIs. For each lesion, we collected all WSIs with the respective diagnosis; some WSIs with multiple diagnoses were placed into multiple datasets. We also collected all WSIs that were normal or unremarkable (n = 1341). Lastly, we also included an ‘all lesions’ category consisting of all WSIs that were not normal, which include WSIs with necrosis, vacuolation, inflammation, and any other lesions (n = 1061). Each category is independently divided into train, validation, and test partitions with quantities of 80%, 10%, and 10% respectively. To form a dataset, we combine the train/validation/test WSIs from the normal category with the train/validation/test WSIs from one of the other categories: necrosis, vacuolation, inflammation, or all lesions. For example, a necrosis dataset would consist of 1073 normal and 91 necrotic training samples, 134 normal and 12 validation samples, and 134 normal and 12 test samples. All quantities are summarized in Table 2. For each dataset (necrosis, vacuolation, inflammation, and all lesions), we randomly generate the train/validation/test splits 8 times to form 8 unique datasets for 8-fold Monte Carlo cross validation.
Preprocessing
Several preprocessing steps are required before obtaining the final feature vectors that can be used for training. First, many of the WSIs contain slices of liver, spleen, and sometimes pancreas. We use a deep learning based in-house segmentation pipeline to segment only liver tissue of each WSI (Fig. 6a). Next, we divide the tissue regions into 256px by 256px tiles with a stride of 256px at 40× magnification. Typically, the number of patches within one WSI is in the tens or hundreds of thousands. The image patches are then passed to a vision transformer which extracts the low-dimensional feature representation of each tile. The lower-dimensional representations enable the simultaneous downstream processing of all tiles of a WSI in GPU memory at once as opposed to in smaller batches. The vision transformers are trained using self-supervised learning approach that uses self-distillation without labels37. We compare the performance of both a feature extractor pretrained on ImageNet38 and a custom feature extractor trained on in-house histology data.
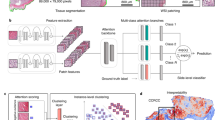
Overview of the pipeline, architectures, and endpoints. (a) Whole slide images are passed through a tissue segmentation network to isolate the desired tissue type. After segmentation, the remaining tissue regions are partitioned into smaller image patches. A pretrained feature extractor encodes the patches into feature representations. (b) During training, the feature vectors are passed through an attention layer to obtain attention scores for each tile indicating importance towards diagnosis. These scores are pooled with the feature vectors and passed to the final classification layer to obtain the slide-level classification. The strongly attended regions are used in the classification pipeline to provide self-guided supervision towards attending to important patches. (c) The inference pipeline reconstructs heatmaps from the attention scores to visualize important regions contributing to the final WSI prediction.
We do not exhaustively screen the training or testing data for the presence of artifacts, though there was an initial quality control step during slide scanning (performed by our in-house expert, T.N.). Instead, we treat the possible presence of artifacts as an intrinsic characteristic of our data. Specifically, stitching artifacts are frequently present and a consequence of the way that pathology images are digitized18,35. Slides are digitized in patches and the patches are composited into a whole slide image. These artifacts are largely imperceptible to the human eye and therefore do not typically impact human review of whole slide images. Stitching and scanning artifacts are present in virtually every whole slide image and can sometimes be picked up by deep learning methods. However, we have shown that our method greatly reduces the detection of these stitching artifacts since our model focusses on more meaningful semantic information in the image.
Algorithm
Overview of weakly-supervised approach with custom embeddings and guided attention
In order to solve the challenges of tissue level classification inherent to digital TOXPATH, we propose a weakly supervised attention-based algorithm heavily inspired by clustering-constrained attention multiple instance learning (CLAM)11. First, each WSI is divided into smaller, easier to process tiles. We then use self -supervised learning techniques to train a feature extractor and generate embeddings for each tile. After we generate the embeddings, we employ a weakly supervised learning method that classifies tissues using only tissue level labels rather than pixel-level annotations. Two main changes that differentiate our approach from CLAM are the usage of a self-supervised vision transformer for feature extraction, and the inclusion of a novel attention mining loss to self-guide the attention layer in place of CLAM’s clustering loss. We find empirically that these changes greatly increase generalizability to a histopathology dataset containing many types of lesions and generate higher quality visualizations. Despite CLAMs improvement on prior methods by using all information on the WSI instead of reducing the entire WSI to a single representative patch, we found that both classification and visualizations generated by CLAM had room for improvement on our in-house datasets.
When adapting CLAM to our application we found two main areas for improvement: first, the usage of a ResNet5039 network as a feature extractor does not extract comprehensive embedding representation of histology patches13. Second, the use of instance-level clustering to refine the feature space is not designed for our TOXPATH dataset as compared to cancer WSIs found in the TCGA dataset. The principal difference between our application and TCGA is the data imbalance in TOXPATH where we cannot always expect both lesioned and non-lesioned tiles to appear in a given slide. In brief, instance-level clustering selects k high and low attended tiles from a WSI and attempts to linearly separate high scores from low scores in order to guide the model’s attention. Therefore, we hypothesized that instance-level clustering results in a similar issue as traditional multiple instance learning, whereby a low number of instances are intended to represent the entire WSI. Additionally, instance-level clustering encourages high contrast between low and high attention regions that is desirable when a lesion is present in a tissue. However, this does not translate well to real world TOXPATH datasets where a majority of tissue sections are normal (contain no lesions) and thus would be expected to generate low contrast attention scores.
Our method may be summarized as a weakly-supervised learning algorithm that trains an attention-based neural network to distinguish the relative importance of regions within a WSI and utilize the information to classify WSIs. The overall pipeline including preprocessing, training, and inference pathways is shown in Fig. 6. First, a tissue segmentation network extracts relevant tissue regions of whole slide images, followed by the partitioning of the regions into smaller image patches (Fig. 6a). A feature extractor network then encodes the image patches into embedding representations. During training, the feature vectors are passed through an attention layer that scores each tile based on diagnostic relevance (Fig. 6b). After pooling the feature vectors and their respective scores, a classification layer generates the final diagnosis. Strongly attended regions are selectively passed through the classification pipeline to provide self-guided supervision that refines attention towards relevant regions. The inference pipeline not only obtains the WSI-level label prediction, but also uses the attention scores to reconstruct heatmaps that highlight significant regions of interest contributing to diagnosis prediction (Fig. 6c).
Details and formulae
The algorithm design is illustrated in Fig. 6b. In brief, a feature extractor reduces each tile to a low dimensional latent representation vector. Each feature vector is then run through a gated attention layer that assigns an attention score to each feature vector. Finally, the model must use the features to classify the WSI as normal or abnormal in two ways: by using all of the features in the WSI and by using a reduced set of only the features corresponding to high attention scores. Forcing the model to classify using the reduced set of high attention features encourages attention to regions that lead to proper classification.
The pretrained feature extractor reduces the dimensionality of each tile to a vector which is the input of the gated attention layer. The Resnet50 (used in CLAM) was trained using fully supervised classification on ImageNet. For the vision transformers, DINO37 was used as the training procedure and we follow the implementation in the original paper exactly. DINO uses self-supervised learning to learn expressive low dimensional representations of images. DINO learns image features by self-distillation, where features derived from random crops of an input image are compared between a teacher and student vision transformer. By comparing large and small image crops, the model learns local-global correspondences in the image. We train our vision transformers using ImageNet (DAM, DGAM) and in house TOXPATH data (CDAM, CDGAM). In both encoder architectures, learned features can be utilized as starting points for downstream tasks such as segmentation and classification.
Using the vision transformer, each tile feature representation is a 1536-dimensional vector. In the gated attention layer, the first fully connected layer \({W}_{1} \in {R}^{1536\times 512}\) further reduces the dimension to a 512-dimensional vector15. For \(n\) patches, we can express the operation as \(H={ZW}_{1}\), where \(Z \in {R}^{n\times 1536}\) is the matrix off input feature representations and \(H \in {R}^{n\times 512}\) is the reduced, learned feature representation after the first layer. In the gated attention layer, there are two parallel layers of the same dimension, \(U \in {R}^{512\times 256}\) and \(V \in {R}^{512\times 256}\), which both take in \(H\) as input. The outputs of these two layers are passed through a \(Sigmoid\) and \(Tanh\) function respectively. The two outputs of the activation layers are multiplied element-wise before passing through another fully connected layer \({W}_{2} \in {R}^{256\times 1}\) that reduces each tile-level feature representation to a single attention score. The \(Softmax\) operation then converts the attention score of each patch into a relative probability. The final matrix \(A \in {R}^{n\times 1}\) that represents the attention scores for all tiles is given by Eq. (1):
The attention scores are then aggregated with the reduced tile feature representation to form a 512-dimensional WSI representation \(S \in {R}^{1\times 512}\) as shown in Eq. (2).
Intuitively, S is a weighted sum of feature vectors where the weights are proportional to the attention scores. S is thus a single vector that describes the entire whole slide image.
The aggregated WSI representation is then passed to a final classification fully connected layer \({W}_{3} \in {R}^{512\times m}\) to obtain the prediction probability vector \(P \in {R}^{1\times m}\) for the \(m\) classes (in our case, m = 2), given by Eq. (3). Classification loss \({L}_{classification}\) is simply the cross-entropy loss when comparing the predicted WSI label to the ground truth slide-label.
Our guided attention mechanism leverages the highly attended scores of positive class instances to provide self-supervision on the attention. The approach is based on the principle that most, if not all, WSIs with lesions still contain instances of normal tissue.
If the attention mechanism is working correctly, then we should be able to determine whether a given WSI should be classified as abnormal by solely looking at the highest scoring tiles. In WSIs with a label indicating the presence of a lesion, the highest scoring tiles should all contain examples of lesions and should have attention scores that are high on the absolute scale. Conversely, in normal WSIs without lesions, we should expect very few high attention tiles, and the highest scoring tiles in that WSI should, by assumption, be lesion free and have attention scores that are lower on the absolute scale and lower than the attention scores of lesioned tiles from the abnormal WSIs. The model should be able to distinguish between normal and abnormal WSIs based the highest scoring tiles from each WSI, since we would expect there to be relatively few high scoring tiles in normal WSIs, and that among these high scoring tiles, their attention scores would still be lower than those found in abnormal WSIs. Therefore, by focusing the model’s attention to these high scoring tiles and considering the diagnostic labels assigned to each WSI, we can suppress high attention values in normal WSIs, and sharpen attention values in abnormal WSIs. We hypothesize that this works especially well in the TOXPATH context where a vast majority of the data belongs to a single, normal class. Whereas CLAM learns to increase contrast between low and high attention regions in all cases, our guided attention method appropriately reduces contrast in normal WSIs. We implement this by sorting the attention scores and selecting only the scores and features associated with the scores for tiles with attention scores above a threshold \(\tau\) (we use \(\tau =0.5\)), given by Eq. (4). We selected the threshold value experimentally to maximize AUC on the validation dataset (see Supplementary Table S2).
We then aggregate the high scores and the associated features and predict the slide-label using the lessened information, as shown in Eq. (5).
The loss \({L}_{mining}\) is an attention mining loss that penalizes highly attending to regions that lead to the wrong classification16. Concretely, it penalizes high probabilities for classes that are not the ground truth class \(c\). The formulation is given in Eq. (6).
Encouraging model attention to a few highly relevant patches has been done in similar works39,40 where the top k scoring features were used to predict the whole slide label. Our method is similar, but by setting a threshold \(\tau\), we dynamically control the number of features that are considered for classification. In normal slides, we would expect that as the model trains few or even zero features could be used for computing mining loss. This allows us to suppress attention scores in normal slides where we know there are no lesions of interest.
Visualization
Visualizations help to increase interpretability of model output and are crucial for integrating these models for WSI diagnostic review workflows. The benefit of assigning attention scores for each patch is that they can be converted to a heatmap representation to highlight the highly attended regions that the model designates as important towards the slide-level label. To achieve this, we save the attention scores for all tiles within a WSI and normalize the range between 0.0 and 1.0. We then map the normalized scores to colors using the jet colormap. Next, we stitch the color tiles together using the coordinates of each associated tile to form a heatmap for the original whole slide image. To improve visualization quality, we can tile the original tissue regions with overlap; for example, we tile the tissue regions into 256px by 256px tiles with a stride of 128px at 40× magnification, which is a 50% overlap. We average the attention scores in the overlapping regions, which improves visualization quality.
Experiments and training details
Our baseline is CLAM with a ResNet50 feature extractor41 that has been pretrained on ImageNet. We compare the baseline CLAM model against 4 different models to assess the impact of our contributions. The 4 models each employ an attention mechanism (CLAM or guided attention) and a vision transformer feature extractor trained on one of two datasets (ImageNet or in house data). Explicitly, we compare the baseline against:
-
(1)
CLAM with a vision transformer feature extractor pretrained with DINO on ImageNet (DAM).
-
(2)
Our guided-attention multiple instance learning method with a vision transformer feature extractor pretrained with DINO on ImageNet (DGAM).
-
(3)
CLAM with a vision transformer feature extractor pretrained with DINO on custom histology data (CDAM).
-
(4)
Our guided-attention multiple instance learning method with a vision transformer feature extractor pretrained with DINO on custom histology data (CDGAM).
We train each model on the 4 datasets. Each dataset was divided into 8 randomized splits of train, validation, and test data. During training, validation loss is calculated at each epoch. After training, the model weights associated with the epoch with lowest validation loss were applied to the test data. The performance on the test data is captured using the area under the receiver operating characteristic (AUROC)42. For each model on each dataset, the mean AUROC is calculated from the 8 splits, along with the standard deviation. The ROC curves are plotted in the results section below, along with representative heatmaps and tiles.
Additionally, since heatmaps are a key algorithmic endpoint of interest, we attempt to quantify heatmap contrast. To do this, we annotated lesions in 20 regions of interest across 3 WSIs with examples of necrosis and computed the contrast in the annotated regions. We define contrast as the ratio between mean heat map pixel intensity in the annotated versus non annotated regions and report the mean contrast and standard deviation among the ROIs.
To enable fair comparison between an ensemble of binary classifiers and a single, multiclass model, we trained a multiclass CDGAM model. Since each WSI can contain multiple kinds of lesions, the multiclass model is allowed to independently predict probabilities for each class. However, since the normal class is by definition mutually exclusive with the lesion classes, particular care was taken to ensure the model makes predictions in this manner. Specifically, if the model predicts the normal class with higher probability than any lesion class, we set the lesion probabilities to zero. Conversely, if the multilabel model predicts any lesions with higher probability than the normal class, we set the normal probability to zero.
For all models, we trained for a maximum of 200 epochs with early stopping based on validation loss over 20 epochs. We use an Adam optimizer43 with a learning rate of 0.0001 and train using a NVidia Quadro P6000. All code is implemented in Python3.6 with PyTorch 1.7.1 as the machine-learning framework. Our vision transformers are standard ViT-S architecture with per token embedding size 384 and patch size 8. For the multiple instance learning, classification heads are configured as in CLAM. We follow the implementation settings in CLAM and direct readers there for additional details.
Data availability
In-house data is currently not publicly available, but requests or inquiries may be addressed to the corresponding author.
Code availability
Our code is based upon the official implementation for CLAM. We release a patch for CLAM that includes our training method, available through Zenodo: https://zenodo.org/records/14617491.
References
Carbonneau, M. A., Cheplygina, V., Granger, E. & Gagnon, G. Multiple instance learning: a survey of problem characteristics and applications. Pattern Recognit. 77, 329–353.
Iizuka, O. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 10, 1504.
Hayakawa, T., Prasath, S., Kawanaka, H., Aronow, B. & Tsuruoka, S. Computational nuclei segmentation methods in digital pathology: a survey. Arch. Comput. Methods Eng. 28, 1–13.
Kurc, T. Segmentation and classification in digital pathology for glioma research: challenges and deep learning approaches. Front. Neurosci. 14, 27.
Pal, A. Deep multiple-instance learning for abnormal cell detection in cervical histopathology images. Comput. Biol. Med. 138, 104890.
Mobadersany, P. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 115, 2970–2979.
Poplin, R. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2, 158–164.
Chen, R. J. et al. Multimodal co-attention transformer for survival prediction in gigapixel whole. Slide Images 4015–4025 (2021).
Hespen, K. M. An anomaly detection approach to identify chronic brain infarcts on MRI. Sci. Rep. 11.
Campanella, G. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309.
Lu, M. Y. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570.
Chen, C. L. An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning. Nat. Commun. 12.
Dehaene, O., Camara, A., Moindrot, O., Lavergne, A. & Courtiol, P. Self-supervision closes the gap between weak and strong supervision in histology. https://doi.org/10.48550/arXiv.2012.03583 (2020).
Ghaffari Laleh N. Benchmarking weakly-supervised deep learning pipelines for whole slide classification in computational pathology.
Ilse, M., Tomczak, J. M. & Welling, M. Attention-based deep multiple instance learning. https://doi.org/10.48550/arXiv.1802.04712 (2018).
Li, K., Wu, Z., Peng, K. C., Ernst, J. & Fu, Y. Tell Me Where to Look: Guided Attention Inference Network. Preprint at https://doi.org/10.48550/arXiv.1802.10171 (2018).
Shao, Z. TransMIL: Transformer based correlated multiple instance learning for whole slide image classification. https://doi.org/10.48550/arXiv.2106.00908 (2021).
Deng, S. Deep learning in digital pathology image analysis: a survey. Front. Med. 14, 470–487.
Jaiswal, A., Babu, A. R., Zadeh, M. Z., Banerjee, D. & Makedon, F. A survey on contrastive self-supervised learning. Technologies 9 (2021).
Chen, R. J. & Krishnan, R. G. Self-supervised vision transformers learn visual concepts in histopathology. https://doi.org/10.48550/arXiv.2203.00585 (2022).
Ciga, O. Overcoming the limitations of patch-based learning to detect cancer in whole slide images. Sci. Rep. 11, 8894.
Azizi, S. et al. Robust and data-efficient generalization of self-supervised machine learning for diagnostic imaging. Nat. Biomed. Eng. 7, 756–779 (2023).
Wang, X. et al. TransPath: transformer-based self-supervised learning for histopathological image classification. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021 (ed de Bruijne, M.) 186–195 (Springer International Publishing, 2021). https://doi.org/10.1007/978-3-030-87237-3_18.
Dosovitskiy, A. An image is worth 16 × 16 words: transformers for image recognition at scale. https://doi.org/10.48550/arXiv.2010.11929 (2020).
Liu, Z. et al. Swin Transformer: Hierarchical Vis. Transformer Using Shifted Windows 10012–10022 (2021).
Deininger, L. A comparative study between vision transformers and CNNs in digital pathology. https://doi.org/10.48550/arXiv.2206.00389.( (2022).
Gao, Z. et al. Instance-based vision transformer for subtyping of papillary renal cell carcinoma in histopathological image. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021 (ed de Bruijne, M.) 299–308 (Springer International Publishing, 2021). https://doi.org/10.1007/978-3-030-87237-3_29
Kang, M., Song, H., Park, S., Yoo, D. & Pereira, S. Benchmarking self-supervised learning on diverse pathology datasets. in IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 3344–3354 (IEEE, 2023). https://doi.org/10.1109/CVPR52729.2023.00326
Hoefling, H. et al. HistoNet: a deep learning-based model of normal histology. Toxicol. Pathol. 49, 784–797 (2021).
Xiang, J. & Zhang, J. Exploring low-rank property in multiple instance learning for whole slide image classification (2022).
Chen, Y. C. & Lu, C. S. RankMix: data augmentation for weakly supervised learning of classifying whole slide images with diverse sizes and imbalanced categories. In IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 23936–23945 (IEEE, Vancouver, 2023). https://doi.org/10.1109/CVPR52729.2023.02292
Bussola, N. et al. A weakly supervised deep learning framework for whole slide classification to facilitate digital pathology in animal study. Chem. Res. Toxicol. 36, 1321–1331 (2023).
Marini, N. et al. Unleashing the potential of digital pathology data by training computer-aided diagnosis models without human annotations. Npj Digit. Med. 5, 1–18 (2022).
Lu, M. Y. et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570 (2021).
Pollatou, A. An automated method for removal of striping artifacts in fluorescent whole-slide microscopy. J. Neurosci. Methods. 341, 108781 (2020).
Tang, W. et al. Multiple Instance Learning Framework with Masked Hard Instance Mining for Whole Slide Image Classification. Preprint at https://doi.org/10.48550/arXiv.2307.15254 (2023).
Caron, M. et al. Emerging Properties in Self-Supervised Vision Transformers 9650–9660 (2021).
Deng, J. et al. ImageNet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255. https://doi.org/10.1109/CVPR.2009.5206848 (2009).
Chikontwe, P., Kim, M., Nam, S. J., Go, H. & Park, S. H. Multiple instance learning with center embeddings for histopathology classification. in Medical Image Computing and Computer Assisted Intervention—MICCAI 2020 (eds Martel, A. L. et al.) 519–528 (Springer International Publishing, 2020). https://doi.org/10.1007/978-3-030-59722-1_50
Zhang, Y. et al. Attention-Challenging Multiple Instance Learning for Whole Slide Image Classification. Preprint at https://doi.org/10.48550/arXiv.2311.07125 (2023).
He, K., Zhang, X., Ren, S. & Sun, J. Deep Residual Learning for Image Recognition 770–778 (2016).
Bradley, A. P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 30, 1145–1159.
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. https://doi.org/10.48550/arXiv.1412.6980 (2014).
Acknowledgements
This work was funded by Genentech.
Author information
Authors and Affiliations
Contributions
J.F. and F.H. conceived the study and designed the experiments. J.F. performed the analyses. P.S. helped with tissue identification. J.F and T.N. curated the in-house datasets. J. F., F.H., R.S., and R.F. reviewed the results and helped with data preparation. P.Z. performed statistical analysis. J.F., P.Z., R.S., R.F., P.S. and F.H. prepared the manuscript. F.H. supervised the research.
Corresponding author
Ethics declarations
Competing interests
All authors were employees of Roche/Genentech at the time the work described in the article was performed. Some of the authors own Roche stock.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zehnder, P., Feng, J., Nguyen, T. et al. Diagnostic classification in toxicologic pathology using attention-guided weak supervision and whole slide image features: a pilot study in rat livers. Sci Rep 15, 4202 (2025). https://doi.org/10.1038/s41598-025-86615-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-86615-6