Abstract
This study aimed to identify potential markers that can predict Parkinson’s disease with mild cognitive impairment (PDMCI). We retrospectively collected general demographic data, clinically relevant scales, plasma samples, and neuroimaging data (T1-weighted magnetic resonance imaging (MRI) data as well as resting-state functional MRI [Rs-fMRI] data) from 173 individuals. Subsequently, based on the aforementioned multimodal indices, a support vector machine was employed to investigate the machine learning (ML) classification of PD patients with normal cognition (PDNC) and PDMCI. The performance of 29 classifiers was assessed based on various combinations of indicators. Results demonstrated that the optimal classifier in the validation set was composed by clinical + Rs-fMRI+ neurofilament light chain, exhibiting a mean Accuracy of 0.762, a mean area under curve of 0.840, a mean sensitivity of 0.745, along with a mean specificity of 0.783. The ML algorithm based on multimodal data demonstrated enhanced discriminative ability between PDNC and PDMCI patients.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is characterized by the prevalent non-motor symptom of cognitive impairment1. Specifically, PD with dementia (PDD) significantly impacts patients’ quality of life and increases mortality rates, adding additional burdens to caregivers2,3. PD with mild cognitive impairment (PDMCI) poses a significantly increased risk of progressing to PDD4. However, studies have also observed that individuals with PDMCI may experience a reversal to normal cognitive function within the context of PD5. Therefore, it is significant to conduct a comprehensive analysis of potential predictors for PDMCI.
Many clinical characteristics are associated with an increased risk of cognitive impairment in PD (PDCI), such as advanced age, low level of education, and male gender. Additionally, higher Hoehn Yahr grade, Rapid Eye Movement (REM) sleep behavior disorder, depression have also been extensively investigated in numerous studies6. The clinical significance of these observed features has been examined in several studies, indicating their potential value in predicting PDCI7. The models established in these studies, however, have not yet been validated in an independent cohort. Several studies have reported associations between PDCI and plasma levels of α-synuclein8, Amyloid-β 1-429, and tau protein10. However, these studies have yielded inconsistent results. Neurofilament light chain protein (NfL) as a potential biomarker for axonal injury and have been found to correlate with cognitive, behavioral, and imaging features of AD as well as predict short-term cognitive decline11. GFAP, recognized as a biomarker for reactive astrocyte hyperplasia, shows promise in the screening of amyloid pathology and predicting future cognitive decline12. Previous studies13 along with our own research14 have also confirmed the strong link between NfL, GFAP, and PDCI. Futhermore, The utilization of neuroimaging markers, such as cortical thickness15. Resting state functional nuclear magnetism (Rs-fMRI) related indicators16 have been identified in multiple studies as potentially valuable for the diagnosis and prediction of PDCI17.
However, one of the major challenges in predicting PDMCI lies in its high heterogeneity18. The application of machine learning (ML) techniques involves using computer-based statistical methods to effectively analyze datasets, allowing clinicians to accurately classify patients based on multiple variables19. Some ML models based solely on resting-state functional magnetic resonance imaging (Rs-fMRI) data20, T1 structural data9,21, and positron emission tomography imaging data22 have shown promising results in differentiating patients with cognitive impairment. However, challenges related with acquiring imaging data seem to impose limitations on sample size and multimodal imaging studies. Some ML studies based on the Parkinson’s Progression Markers Initiative (PPMI) databases also integrate plasma and cerebrospinal fluid (CSF) markers, including alpha-Synuclein, Aβ protein, tau protein, tau phosphate 181, and genetic markers10,23. However, the higher level of education in the PPMI dataset may affect the generalizability of findings to individuals with lower education levels. Furthermore, the integration of plasma NfL and GFAP measures into ML models to assess their impact on model performance has not yet been explored. Therefore, it is highly anticipated that ML can be employed to evaluate the role of NfL and GFAP in identifying PDMCI.
The present study integrated clinical, plasma, and imaging indicators, including T1 data and Rs-fMRI data, to construct PDMCI models with different combinations of indicator using ML and the effects of these models were effects of these models were compared. An overview of the framework employed for multi-indicator machine learning classification, including multi-indicator collecting, feature selection, model construction, and model evaluation in Fig. 1. It was aimed to investigate which specific combination of indicators plays a more significant role in recognizing PDMCI.
Results
Analysis of clinical data and plasma features
A total of 173 participants completed clinical data collection, plasma NfL, and GFAP level measurements, as well as imaging assessments (including T1 and Rs-fMRI). The Healthy control (HC) group consisted of 32 individuals, while the PD with normal cognition (PDNC) and PDMCI groups included 69 and 72 participants, respectively. Table 1 presents the clinical data and plasma analysis results for all three patient groups. Compared to the HC and PDNC groups, PDMCI group was elder, and showed lower levels of education and assessment utilizing the Montreal Cognitive Assessment (MoCA) scores, as well as higher Hoehn Yahr scale, Unified Parkinson’s Rating Scale Part III (UPDRSIII), rapid eye movement sleep behavior disorder screening questionnaire (RBDSQ) scores, Hamilton anxiety scale (HAMA) scores, Hamilton depression scale (HAMD) scores, Pittsburgh Sleep Quality Index (PSQI) scores, and freezing of gait questionnaire (FOGQ) scores with statistical significance. Additionally, plasma NfL and GFAP levels were significantly elevated. There were no significant disparities observed in terms of gender distribution, levodopa equivalent daily dose (LEDD), and disease duration among the groups.
Analysis of neuroimaging features
T1 features
Significant differences were observed in the cortical thickness (Cth) values of 10 brain regions and the volume of 7 subcortical structures between the PDNC and PDMCI groups. These differences were determined through independent sample T-tests conducted on the extracted data after Z-score transformation during pre-processing (Table 2).
Rs-fMRI features
The normally distributed Rs-fMRI data underwent Z-value conversion during data preprocessing. Independent sample T-tests were employed to perform statistical analysis on Amplitude of low-frequency fluctuation (ALFF), Fractional ALFF (fALFF), Regional homogeneity (ReHo), Functional connectivity density (FCD) values extracted from 116 brain regions, and 6670 FC values (The FC matrix is non-directional, therefore it is advisable to consider only half of its values). The results demonstrated the differences in the ALFF values of 10 brain regions, fALFF values of 1 brain region, FCD values of 2 brain regions, ReHo values of 9 brain regions, and Functional connectivity (FC) values of 7 brain regions between the PDNC and PDMCI groups with statistical significance (Table 3).
Evaluation outcomes of the SVM classifier
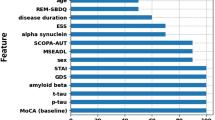
The result of the difference analysis encompassed a total of 9 clinical features, 2 plasma features, 17 T1 structural features, and 29 Rs-fMRI features. Feature selection was performed using RF based on their importance, resulting in the identification of the top 2 features for inclusion in model training. Figure 2 visually represents the significance of each indicator. The study identified two clinical features (FOGQ and education), two plasma markers (NfL and GFAP), two T1 structural features (Cth value of the left postcentral gyrus and volume of the right nucleus accumbens), as well as two Rs-fMRI features (FC of the right Orbitofrontal gyrus and Vermis_4,5; together with FC value of the left inferiorfrontal_Oper and left lingual).
a denotes the ranking of clinical features in terms of their importance, b represents the ranking of T1 structural features based on their significance, and c signifies the importance ranking of Rs-fMRI features. ALFF low-frequency amplitude, fALFF fractional low-frequency amplitude, FC Functional connection, FCD Functional connection density, FOGQ Freezing of Gait questionnaire, HAMA Hamilton Anxiety Scale, HAMD Hamilton Depression Scale, HY Hoehn Yahr, lh left hemisphere, PSQI Pittsburgh Sleep Quality Index, RBDSQ Rapid Eye Movement Sleep Behavior Disorder Scale, ReHo local consistency, rh right hemisphere, UPDRSIII Unified Parkinson’s Rating Scale Part III.
The performance of 29 classifiers was assessed to determine the optimal combination of indicators that can effectively distinguish PDNC from PDMCI (Table 4). The results demonstrated that the optimal classifier in the validation set included clinical data combined with Rs-fMRI and NfL biomarkers, mean accuracy (ACC) of 0.762, a mean area under curve (AUC) of 0.840, a mean sensitivity of 0.745, as well as a mean specificity of 0.783 (Fig. 3a). Subsequently, clinical features combined with Rs-fMRI and GFAP markers also achieved favorable performance (mean ACC: 0.759, mean AUC: 0.834, mean sensitivity: 0.783, mean specificity: 0.736, Fig. 3b). Furthermore, good performance was observed when integrating clinical variables with GFAP measurements (Fig. 3c).
Features impacting PDNC and PDMCI classification
The best-performing classifier, incorporating clinical + Rs-fMRI + NfL markers, was assessed for its contribution to classifier prediction by calculating the average absolute SHapley Additive exPlanation (SHAP) value for each feature. The global feature importance chart in Fig. 4 illustrates the descending order of importance. The findings indicated that within the classifiers established by clinical + Rs-fMRI + NfL indicators, plasma NfL showed the highest global importance, followed by education, while the FC value of the left inferiorfrontal_Oper and left lingual demonstrated the lowest significance.
The ordinate of the graph is determined by arranging features in descending order of importance, while the horizontal coordinate represents the average absolute SHAP value of the feature. Cth cortical thickness, FOGQ Freezing of Gait questionnaire, GFAP glial fibrillary acidic protein, lh left hemisphere, NfL plasma neurofilament light chain, PDNC Parkinson’s disease with normal cognition, PDMCI Parkinson’s disease with mild cognitive impairment, rh right hemisphere.
The SHAP value of each feature for every sample was visualized by plotting a scatter on the SHAP summary graph. As depicted in Fig. 5, the relationship between the magnitude of the feature value and the classifier’s predicted impact could be observed through color, enabling visualization of its feature value distribution. The findings revealed that elevated levels of NfL, and FOGQ scores, as well as reduced education level, FC of the right Orbitofrontal gyrus and Vermis_4,5; FC value of the left inferiorfrontal_Oper and left lingual were associated with an increased risk of PDMCI.
The ordinate is based on the features arranged in descending order of importance, while the horizontal coordinate represents the SHAP value of the feature. Positive values indicate membership in the PDMCI group, while negative values indicate membership in the PDNC group. The scatter plots in the figure depict SHAP values for each feature of every sample. The color of the scatter and the red-blue color bar on the right indicate the magnitude of eigenvalues for each sample, with red denoting higher eigenvalues and blue denoting lower eigenvalues. For instance, the risk indicator for major PDMCI, NfL, exhibits a pronounced right-skewed distribution with a cluster of high values, indicating an increased risk of PDMCI associated with elevated NfL levels. Cth Cortical thickness, FOGQ Frozen gait questionnaire, GFAP glial fibrillary acidic protein, NfL Plasma neurofilament light chain, PDNC Parkinson’s disease with normal cognitive, PDMCI Parkinson’s disease with mild cognitive impairment.
The SHAP dependency graphs in Fig. 6 illustrate the crucial predictors, enabling the identification of both the direction and threshold at which risk escalates. For example, the levels of NfL reached a threshold of ≥13 pg/mL (depicted on the X-axis in Fig. 6a). As these levels increased, SHAP values (represented on the Y-axis in Fig. 6a) exhibited a progressively positive trend, indicating a significant predictive capability for later diagnosis of PDMCI in PD patients. The thresholds for the five main predictors were indicated by red dashed lines in the figure : NfL ≥ 13 pg/mL; FOGQ ≥ 9 points; Education ≤6 years; FC of the right Orbitofrontal gyrus and Vermis_4,5 ≤ 0.255; and volume of right nucleus accumbens ≤0.19.
The graphs illustrate the threshold at which an elevated risk occurs for each predicted value. The X-axis represents the value of the predictor, while the Y-axis represents the SHAP value of the predictor. The predicted value on the X-axis directly influences the corresponding SHAP value on the Y-axis. Points on the X-axis that correspond to SHAP values above 0 indicate thresholds associated with increased risk. Each dot in the diagram represents each subject. Values located to the right of the red dashed lines indicate an elevated risk of PDMCI (b, c). Conversely, values situated to the left of the red dotted line also suggest an increased risk of PDMCI (a, d and e). FOGQ Frozen gait questionnaire, GFAP glial fibrillary acidic protein, NfL Plasma neurofilament light chain, PDMCI Parkinson’s disease with mild cognitive impairment.
Discussion
In this study, we collected a comprehensive and multimodal dataset including general demographic information, clinical neuropsychological assessments, plasma indicators (NfL and GFAP), T1 structural data, and Rs-fMRI data. Our analysis, using ML algorithms, revealed that among the 29 classifiers tested, the one integrating multi-modal indicators—comprising clinical assessment, Rs-fMRI indicator, and NfL—demonstrated superior performance.
The incorporation of clinical indicators obviously enhanced the accuracy and efficacy of the 29 established classifiers. Our findings based on feature importance indicated that FOGQ and education provided the most valuable insights for distinguishing PDNC from PDMCI, surpassing other clinical metrics. When analyzing the top-performing classifier 1 using SHAP, FOGQ turned out to be significant important (ranking second). This underscored a robust link of FOGQ to cognitive impairment within PD. Cholinergic dysfunction may serve as a shared underlying factor contributing to both cognitive impairment in PD and the occurrence of FOG24,25. Moreover, structural and functional imaging studies have implicated the frontal lobe—an executive brain region—in PD-related FOG26 and cognitive dysfunction17. Additionally, research by Monaghan et al. revealed that patients with FOG exhibited more severe disease progression compared to those without FOG, along with more pronounced cognitive impairment27, indicating that FOG may exacerbate motor symptoms and contribute to cognitive decline. The relationship between education and cognitive function has been well-established. Numerous studies have demonstrated a positive correlation between the duration of formal education and cognitive function during adulthood, as well as a reduced risk of dementia later in life28. Our findings further validated that a higher level of education might serve as an indicator of cognitive reserve, providing protection against cognitive decline in patients with PD.
While clinical indicators can improve the classifier’s performance to some degree, relying solely on them cannot yield optimal results. Among the 29 classifiers we established, the top ten performers all integrated plasma indicators. NfL demonstrated the highest global feature importance in our analysis, emphasizing the significant role of plasma in distinguishing PDNC from PDMCI. Our study incorporated novel plasma markers of cognitive impairment, namely NfL and GFAP, which have garnered attention in recent years. Our findings were in accordance with other studies conducted in various countries and regions, indicating a significant elevation of plasma NfL levels among patients with PDMCI14,29,30. Existing evidence suggests that elevated NfL levels among PD patients with cognitive impairment may be attributed to augmented white matter lesions or alterations in gray matter structure31,32,33. While few studies have explored the role of GFAP in cognitive dysfunction associated with PD, most research has focused on Alzheimer’s disease (AD). Utilizing the ultrasensitive Simoa platform, our study revealed obviously higher levels of plasma GFAP in the PDMCI group in contrast with both the HC and PDNC groups. These findings were consistent with other studies supporting the link between GFAP and cognitive dysfunction within PD13,34. However, Oeckl et al. found that GFAP concentration in the PDNC group was similar to that in the PDD group35. Under pathological conditions, an abnormal increase in GFAP expression might indicate the activation of astrocytes36, which plays a crucial role in the development of cognitive impairment in PD37,38. In conclusion, NfL together with GFAP might hold promise as potential peripheral biomarkers for assessing cognitive function in individuals with PD.
Another crucial aspect for identifying PDMCI is neuroimaging indicators. Previous studies, including our own research, have shown extensive cortical atrophy and alterations in functional connectivity among PD individuals with cognitive impairment17. However, to date, no studies have investigated the ability to identify PDMCI using ML algorithms that directly compare or combine T1 and Rs-fMRI data. Our feature selection results, based on the importance of features using RF (Fig. 2b), reveal that the top two Rs-fMRI indicators are the FC of the right Orbitofrontal gyrus and Vermis_4,5; together with FC value of the left inferiorfrontal_Oper and left lingual. The prefrontal cortex is pivotal in executive function39, while damage to the cerebellum can lead to executive and visuospatial disturbances40. Certain studies propose that increased cerebellar metabolism41 and enhanced functional connectivity linked to the cerebellum42 may represent compensatory mechanisms aimed at preserving cognitive function in individuals with PDCI. We observed reduced FC in the right Orbitofrontal gyrus and vermis, as well as between other brain regions, in patients with PDMCI, compared to controls and PDNC individuals, and plays a prominent role in distinguishing PDNC from PDMCI. Therefore, we speculate that the “prefrontal cortex-cerebellar circuit” might play a compensatory role to some extent in the decline of cognitive function in PD, and when degeneration occurs within this circuit’s functional connectivity, it could be an important mechanism for the progression of cognitive dysfunction. The SHAP summary and dependency maps suggest that lower FC value of the left inferiorfrontal_Oper and left lingual may increase the risk of PDMCI. The lingual gyrus is associated with visuospatial dysfunction43,44. The presence of visuospatial dysfunction is a prominent cognitive manifestation observed in PD45. Some studies have suggested that the visuospatial ability of individuals with PD is associated with the dorsolateral prefrontal circuit and the lateral orbitofrontal circuit45. However, frontal executive dysfunction alone does not fully account for the observed visuospatial deficits in PD. Additional research has indicated that visuospatial impairments in PD often involve posterior brain regions, which may play a dominant role in visuospatial abilities46. The above results further validated the link between cognitive impairment among PD patients and the involvement of both the central executive and visual networks47.
While our findings suggest that the Rs-fMRI indicator outperforms the T1 structural indicator in identifying PDMCI, the results from T1 structural indicator provide significant insights as well. The primary contributors to the performance of our model are predominantly the Cth of the left postcentral gyrus and the volume of the right nucleus accumbens in T1 structural features. The postcentral gyrus serves as a cortical hub for somatosensory processing, integrating both the dorsal attention network and the default network48. These interconnected networks are closely associated with cognitive functioning47. Notably, we found that the volume of the nucleus accumbens is more significant than other subcortical volume features. Previous studies have also reported progressive atrophy of the nucleus accumbens in PDMCI patients49. As a prominent component of the ventral striatum50, the nucleus accumbens primarily projects into the frontal cortex and limbic pathways, serving as an indicative marker for overall dysfunction within the mesocorticolimbic network51. Moreover, there is evidence suggesting that atrophy of the nucleus accumbens in PD patients may lead to acetylcholinesterase deficiency, subsequently contributing to the development of PDD52.
The SVM algorithm employed in this study is a widely utilized ML modeling technique within the realm of neuroimaging. By identifying a hyperplane in a high-dimensional space based on two types of data, it effectively segregates the remaining data into two distinct categories. Due to its unique principles and algorithms53, SVM exhibits certain advantages when analyzing small sample sizes. By employing ML techniques, our study demonstrated that using a single modality (clinical/T1 structure/Rs-fMRI/plasma) might be insufficient for distinguishing PDNC from PDMCI patients. The integration of all indicators, however, does not represent an optimal model. The classifier combining clinical, Rs-fMRI, and NfL indicators demonstrated superior accuracy, classification efficacy, specificity, and sensitivity in discriminating PDNC from PDMCI patients. These findings provided robust evidence for further exploration of potential predictors for PDD.
Nevertheless, some flaws also exist in the present study. Firstly, we refrained from exploring the potential of deep learning algorithms and unsupervised machine learning to enhance predictive performance due to the absence of clear clinical guidance information and the intricacy in interpreting and comprehending the models. Secondly, the sample size was relatively small for neuropsychological evaluation, blood testing, and multimodal MRI data collection. Lastly, our findings were not validated by external or longitudinal data. Thus, longitudinal studies using larger multicenter cohorts are essential for identifying potential markers of PDMCI in the future.
In conclusion, the multi-modal ML algorithm demonstrated improved discriminative ability between PDNC and PDMCI patients. The incorporation of clinical, Rs-fMRI, and NfL indicators resulted in superior accuracy, classification efficacy, specificity, and sensitivity in distinguishing PDNC from PDMCI.
Methods
Participants
This retrospective study was carried out at the Department of Neurology, the First Affiliated Hospital of Kunming Medical University, from February 2022 to December 2023, including both inpatients and outpatients. Participants were categorized into three groups: HC, PDNC, and PDMCI. The diagnosis of PD patients followed the diagnostic criteria established by the International Parkinson’s Disease and Movement Disorders Society (MDS) in 201554. The assessment of PDMCI was conducted in accordance with the Type I diagnostic criteria for PDMCI published by MDS in 201255. The PDNC group comprised PD patients not meeting the diagnostic criteria for PDMCI55 and PDD56. Family members of both inpatient and outpatient patients, as well as individuals examined as HC group, exhibited no cognitive decline based on assessment utilizing the MoCA.
Exclusion criteria were as follows: (1) Patients with atypical/secondary symptoms linked to PD, such as progressive supranuclear palsy, multiple system atrophy, corticobasal ganglion degeneration, vascular PD, as well as antipsychotic drug-induced Parkinsonism; (2) Patients with cognitive decline attributed to other underlying conditions; (3) Patients with a history of mental illnesses such as depression or anxiety disorders; (4) Patients with coexisting medical conditions significantly impacting cognitive or other scales assessments, such as aphasia, delirium, epilepsy, or severe diseases; (5) Patients with prior intracranial surgery for PD management; (6) Patients who were left-handed; (7) Patients who were unable to cooperate in completing scale assessments.
The study was approved by the ethics committee of the First Affiliated Hospital of Kunming Medical University (2022-L-46). All participants provided informed consent, and the study adhered to the principles outlined in the World Medical Association Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
Clinical scale evaluation
Demographic information, including age, gender, and educational background (with years of formal education measured), was collected for all eligible participants. For PD patients, data on disease duration, medication regimen (with daily levodopa dosage calculated), and motor function were assessed using the UPDRSIII as well as the Hoehn and Yahr scale. Mood assessment for PD patients consisted of the HAMA and HAMD, while cognitive status was assessed using the MoCA. The RBDSQ was utilized to assess symptoms of REM sleep behavior disorder57. The PSQI was employed to evaluate sleep quality in PD patients, and freezing of gait (FOG) was evaluated using the FOGQ58.
Regarding cognitive status assessment, each patient underwent evaluation independently by two associate chief physicians or above to ensure diagnostic reliability and minimize potential selection bias in the study. Furthermore, all scale assessments were carefully reviewed by two attending physicians who received standardized training to ensure accurate measurements.
Collection and analysis of blood samples
Venous blood samples (5 mL) were collected from each subject, and within 1 h of collection, the sample was centrifuged at a force of 2500 g for a duration of 10 min. Subsequently, the plasma was separated and stored in a polypropylene tube at −80 °C for future use. Analysis was performed through the Simoa HD-1 analyzer (Quanterix, MA, USA) following the instruction provided by the manufacture. The samples underwent a single thawing process for NfL and GFAP measurements. Manual dilution of the sample was not performed; instead, it was automatically diluted fourfold by the detector following the recommended protocol provided by the manufacturer. The coefficient of variation for multiple measurements was determined to be 4.2%. In each test, two quality control samples were incorporated, featured by pre-provided concentrations of high and low NfL and GFAP that fell within the anticipated range. The above experiments were conducted by research assistants who remained blind to the patients’ diagnoses.
Acquisition of neuroimaging data
The MRI data were acquired using a 3T full-body scanner located in the radiology department of a hospital (Discovery 750w, GE Healthcare, Fairfield, USA). Standard head coils were used for both transmit and receive functions. Prior to the scan, participants were informed about the requirements for the scanning. They reclined in a relaxed state without engaging in sleep or cognitive activities. Earplugs were worn by all subjects to attenuate external auditory stimuli, while their heads remained immobilized to minimize potential motion artifacts.
The following parameters were used to acquire a three-dimensional T1-weighted fast gradient echo sagittal image with magnetization preparation: 156th floor, layer thickness: 1 mm, no spacer layer, voxel size: 1 × 1 × 1 mm, repetition time 1.900 ms, echo time: 2.0 ms, inversion time: 450 ms, flip angle: 12°, field of view: 256 mm, matrix: 256 × 256. Rs-fMRI images were obtained via ultrasonic planar imaging: 36 layers, layer thickness: 3 mm, no spacer layers, voxel size: 3.5 × 3.5 × 4 mm, volume: 240, repetition time: 2000 ms, echo time: 30 ms, flip angle: 90°, field of view: 224 mm, matrix: 64 × 64.
Preprocessing of neuroimaging data
MRI scans were carefully performed to exclude any significant medical irregularities and to ensure that the images were of high quality. For the analysis of Cth, we used FreeSurfer 6.0.0, (https://surfer.nmr.mgh.harvard.edu/, Ubuntu Linux) along with the Surface-based morphometry (SBM) method for processing T1-weighted images.
The analysis involved several main steps: Firstly, the three-dimensional T1 image was converted into a four-dimensional format using MRIcron software for subsequent processing and analysis. The data were then imported into FreeSurfer 6.0.0 on Linux Ubuntu for automated processing, which included head motion correction, removal of extraneous brain structures (such as skulls), automatic conversion to Talairach space, segmentation of subcortical structures, gray normalization, stitching of gray matter boundaries, automatic topological correction, deformation of brain tissue surfaces, expansion of surfaces after cortical image generation, as well as conversion to spherical distribution templates.
Cth of each brain region was calculated following Gaussian smoothing with a kernel (half maximum width 10 mm). Cth was defined as the distance between the boundary of gray matter/white matter and the corresponding top surface of the brain. The reconstructed datasets underwent thorough examination to ensure the accuracy of registration, cranial anatomy, segmentation, as well as cortical surface reconstruction.
The analysis of Rs-fMRI images was conducted using the MATLAB2013b platform (https://www.mathworks.com/products/matlab.html). Preprocessing was carried out with the help of the Brain Imaging Data Processing and Analysis (DPABI) toolbox (version 4.5, http://www.rfmri.org/dpabi).
Initially, the first ten time points were excluded to minimize potential artifacts arising from the scanner’s initial setup and the subjects’ acclimatization to the scanning surroundings. The remaining images underwent slice time correction, utilizing the middle layer as a reference, followed by realignment to eliminate head movements. Images with head displacement more than 2 mm or angular rotation exceeding 2° along the x, y, and z axes were excluded to enhance the robustness of functional connection analysis against head motion artifacts. Subsequently, T1-weighted anatomical images were aligned with the average functional images, and the structural images were segmented into gray matter, white matter, and CSF using the diffeomorphic anatomical registration through exponentiated lie algebra (DARTEL) template. The data were normalized to the DARTEL template in Montreal Neurological Institute (MNI) space to enhance anatomical precision across participants. The normalized Rs-fMRI data were resampled into 3 × 3 × 3 mm3 voxels based on spatial smoothing with a Gaussian kernel of half maximum width of 6 mm. The low-frequency component was removed through de-linear drift correction, and physiological high-frequency noise was filtered out using a 0.01–0.08 Hz bandpass filter. Interference covariates, such as signals from white matter and CSF, were regressed along with Friston-24 head motion parameters, including 6 parameters measuring head motion alongside their historical effects, as well as 12 corresponding square terms.
Compute the indicator of the Rs-fMRI
(1) ALFF: ALFF reflected the magnitude of spontaneous oscillatory functional activity in the brain, specifically referring to the intensity of neural activity. During the preprocessing of Rs-fMRI data, no filtering processing was applied to calculate ALFF. Following fast Fourier processing, the time domain signal in each voxel’s time series was transformed into the frequency domain. Subsequently, the power spectrum and its square root were computed, with the average calculated within the range of 0.01 to 0.08 Hz. The zALFF map of the entire brain was derived through Fisher z transformation and aligned with the anatomical automatic labeling (AAL) brain atlas. The AAL atlas segmented the brain into 116 distinct regions, enabling the acquisition of zALFF values for each of these regions.
(2) fALFF: Similar to ALFF, fALFF also reflected the magnitude of spontaneous neural activity in the brain. The procedure for calculating fALFF was identical to that of ALFF, except that whole-brain fALFF was obtained by computing the ratio between the average power spectrum and the sum of amplitudes across the entire frequency domain.
(3) FCD: FCD primarily reflected the network connectivity characteristics of a specific voxel across various scales, evaluating the significance of a brain region by quantifying its connections with other brain regions. For FCD calculation, the frequency domain of the filter was set between 0.01–0.1 Hz during data preprocessing. Once the FCD value was calculated, it would undergo a smoothing process with a half maximum width of 8 mm. The FCD values of 116 brain regions were initially computed on the basis of the AAL brain Atlas.
(4) ReHo: ReHo measured the extent to which the intrinsic neuronal activity of a specific brain region aligned with that of its neighboring regions, focusing on local brain connections. It indicated the consistency between a voxel and its adjacent voxels during the scanning period. The ReHo value was calculated before the smoothing process (with a half maximum width of 6 mm). The ReHo value was obtained by computing the temporal correlation between a voxel and its 26 neighboring voxels. Subsequently, segmentation based on the AAL brain map allowed for obtaining ReHo values for 116 brain regions. Finally, Fisher z transformation was applied to obtain zReHo values for these 116 brain regions.
(5) FC: FC captured spatial-temporal correlations among neural activities across distinct brain regions. Pearson correlation coefficients were computed between the average time series across 116 brain regions, based on the AAL brain map, using pre-processed data. Subsequently, the resulting connectivity map was transformed into a z map through Fisher z transform for subsequent statistical analysis.
ML
The ML algorithm was implemented using Python 3.9, a platform that has shown successful application in various biomedical research endeavors. The key steps included feature extraction, feature selection, model establishment, and evaluation, as well as model interpretation.
Extraction of neuroimaging features
As mentioned above, the preprocessing of T1 involved SBM measurement analysis, which was conducted using the FreeSurfer software. FreeSurfer offered a precise method for measuring the thickness of the entire cerebral cortex and provided detailed segmentation of brain regions based on its Desikan-Killiany-Tourville partitioning approach. This partitioning technique divided the bilateral cerebral cortex into 68 brain regions and 12 subcortical structures, including the bilateral pallidum, caudate nucleus, putamen, hippocampus, nucleus accumbens, and amygdala. We extracted the Cth of 68 brain regions and the volume of 12 subcortical structures as characteristic indicators for T1. On the MATLAB2013b platform, we extracted the ALFF, fALFF, FCD, and ReHo values of 116 brain regions obtained after preprocessing. Additionally, we calculated the FC values between each pair of these 116 brain regions, resulting in a total of 6670 FC indicators.
Feature selection
Feature screening was a crucial process in ML aimed at identifying variables with predictive value. In this study, we adopted a combination of differential analysis and feature importance analysis for effective feature screening. Clinical and plasma features were analyzed using SPSS 25.0 (IBM, Chicago, IL, USA). The Kolmogorov-Smirnov method was employed to assess the normality of continuous variables, and the independent sample T-test was adopted for data with normal distribution; otherwise, the Mann-Whitney test was applied. Chi-square tests were conducted for counting variables. In Python 3.9, independent sample T-tests were performed to analyze T1 structural features and Rs-fMRI features. Features with significant differences between groups (P < 0.05) were selected, and to prevent overfitting, FC values below P < 0.001 were considered. The scikit-learn library (http://scikit-learn.org)59 in Python was then utilized for classification based on information gain (IG), where random forest (RF) was used to determine the significance of features and identify the most relevant ones22. IG was a feature evaluation method based on entropy, which measured the amount of information provided by a feature during the classification process, reflecting its usefulness. RF was used to assess importance by calculating the increase in prediction error caused by each feature.
Model establishment and evaluation
The “train_test_split” function was employed to randomly partition 70% of the PD patients as the training set and allocate 30% as the validation set. Prior to model training, min-max normalization and mean regularization were applied to enhance the data structure for training. This step was aimed to mitigate the impact of variations in data distribution regions on convergence speed and prediction accuracy.
We employed a linear kernel SVM for model training, a widely adopted ML technique. Its fundamental principle involved identifying an optimal hyperplane or decision boundary that maximized the margin between data points of different categories, by minimizing misclassification and achieving high prediction accuracy in classification problems53. The SVM classifier was validated, and the best hyperparameters were selected using fivefold cross-validation, considering the sample size and preventing overfitting. The stability of the results was assessed by repeating the entire process, including data set partitioning, feature normalization, regularization, SVMs, and 5-fold cross-validation, for a total of 100 times. Subsequently, the model’s mean ACC, mean AUC, mean sensitivity, and mean specificity were evaluated.
Model interpretation
SHAP values were used to assess the significance of the features incorporated in the model. Originating from cooperative game theory, SHAP values were interpreted within ML as a means to figure out the variables’ contributions to predictions60. These values served as a valuable tool for comprehending the decision-making process of ML models. By analyzing the individual impact of each feature on prediction outcomes, SHAP values provided deeper insights into the underlying reasoning of the model. The SHAP library in Python 3.9 was utilized to generate three distinct SHAP plots: the global feature importance plot, the SHAP summary plot, and the SHAP dependency plot, offering complementary interpretations of the acquired prediction model.
Data availability
The data supporting the conclusions of this article are included within the article and its additional files. Additional data are available from the corresponding author upon request.
References
Aarsland, D. et al. Risk of dementia in Parkinson’s disease: a community-based, prospective study. Neurology 56, 730–736 (2001).
McMahon, L., Blake, C. & Lennon, O. A systematic review and meta-analysis of respiratory dysfunction in Parkinson’s disease. Eur. J. Neurol. 30, 1481–1504 (2023).
Leroi, I., McDonald, K., Pantula, H. & Harbishettar, V. Cognitive impairment in Parkinson’s disease: impact on quality of life, disability, and caregiver burden. J. Geriatr. psychiatry Neurol. 25, 208–214 (2012).
Cosgrove, J., Alty, J. E. & Jamieson, S. Cognitive impairment in Parkinson’s disease. Postgrad. Med. J. 91, 212–220 (2015).
Pedersen, K. F., Larsen, J. P., Tysnes, O. B. & Alves, G. Natural course of mild cognitive impairment in Parkinson disease: a 5-year population-based study. Neurology 88, 767–774 (2017).
Hogue, O., Fernandez, H. H. & Floden, D. P. Predicting early cognitive decline in newly-diagnosed Parkinson’s patients: a practical model. Parkinsonism Relat. Disord. 56, 70–75 (2018).
Chung, S. J. et al. Factor analysis-derived cognitive profile predicting early dementia conversion in PD. Neurology 95, e1650–e1659 (2020).
McFall, G. P. et al. Identifying key multi-modal predictors of incipient dementia in Parkinson’s disease: a machine learning analysis and Tree SHAP interpretation. Front. Aging Neurosci. 15, 1124232 (2023).
Shin, N.-Y. et al. Cortical thickness from MRI to predict conversion from mild cognitive impairment to dementia in Parkinson disease: a machine learning–based model. Radiology 300, 390–399 (2021).
Almgren, H. et al. Machine learning-based prediction of longitudinal cognitive decline in early Parkinson’s disease using multimodal features. Sci. Rep. 13, 13193 (2023).
Kern, D. et al. Serum NfL in Alzheimer dementia: results of the Prospective Dementia Registry Austria. Medicina 58, 433 (2022).
Yang, Z. et al. Clinical and biological relevance of glial fibrillary acidic protein in Alzheimer’s disease. Alzheimers Res Ther. 15, 190 (2023).
Tang, Y. et al. Plasma GFAP in Parkinson’s disease with cognitive impairment and its potential to predict conversion to dementia. NPJ Parkinsons Dis. 9, 23 (2023).
Zhu, Y. et al. Association between plasma neurofilament light chain levels and cognitive function in patients with Parkinson’s disease. J. Neuroimmunol. 358, 577662 (2021).
Pagonabarraga, J. et al. Pattern of regional cortical thinning associated with cognitive deterioration in Parkinson’s disease. PLoS One 8, e54980 (2013).
van den Heuvel, M. P. & Hulshoff Pol, H. E. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur. Neuropsychopharmacol. 20, 519–534 (2010).
Zhu, Y. et al. Cortical atrophy is associated with cognitive impairment in Parkinson’s disease: a combined analysis of cortical thickness and functional connectivity. Brain Imaging Behav. 16, 2586–2600 (2022).
Hely, M. A., Reid, W. G., Adena, M. A., Halliday, G. M. & Morris, J. G. The Sydney multicenter study of Parkinson’s disease: the inevitability of dementia at 20 years. Mov. Disord. 23, 837–844 (2008).
Landolfi, A. et al. Machine learning approaches in Parkinson’s disease. Curr. Med. Chem. 28, 6548–6568 (2021).
Abós, A. et al. Discriminating cognitive status in Parkinson’s disease through functional connectomics and machine learning. Sci. Rep. 7, 45347 (2017).
Zhang, J. et al. An SBM-based machine learning model for identifying mild cognitive impairment in patients with Parkinson’s disease. J. Neurol. Sci. 418, 117077 (2020).
Amboni, M. et al. Machine learning can predict mild cognitive impairment in Parkinson’s disease. Front. Neurol. 13, 1010147 (2022).
Harvey, J. et al. Machine learning-based prediction of cognitive outcomes in de novo Parkinson’s disease. npj Parkinson’s Dis. 8, 150 (2022).
Morris, R. et al. Overview of the cholinergic contribution to gait, balance and falls in Parkinson’s disease. Parkinson. Relat. Disord. 63, 20–30 (2019).
Bohnen, N. I. et al. Cholinergic system changes of falls and freezing of gait in Parkinson’s disease. Ann. Neurol. 85, 538–549 (2019).
Bosch, T. J., Barsainya, R., Ridder, A., Santosh, K. C. & Singh, A. Interval timing and midfrontal delta oscillations are impaired in Parkinson’s disease patients with freezing of gait. J. Neurol. 269, 2599–2609 (2022).
Monaghan, A. S. et al. Cognition and freezing of gait in Parkinson’s disease: a systematic review and meta-analysis. Neurosci. Biobehav. Rev. 147, 105068 (2023).
Lövdén, M., Fratiglioni, L., Glymour, M. M., Lindenberger, U. & Tucker-Drob, E. M. Education and Cognitive Functioning Across the Life Span. Psychol. Sci. Public Interest 21, 6–41 (2020).
Lin, C.-H. et al. Blood NfL. Neurology 93, e1104–e1111 (2019).
Arai, H. et al. Epitope analysis of senile plaque components in the hippocampus of patients with Parkinson’s disease. Neurology 42, 1315–1322 (1992).
Mattsson, N. CSF biomarkers in neurodegenerative diseases. Clin. Chem. Lab. Med. 49, 345–352 (2011).
Bohnen, N. I. & Albin, R. L. White matter lesions in Parkinson disease. Nat. Rev. Neurol. 7, 229–236 (2011).
Xu, Y., Yang, J., Hu, X. & Shang, H. Voxel-based meta-analysis of gray matter volume reductions associated with cognitive impairment in Parkinson’s disease. J. Neurol. 263, 1178–1187 (2016).
Liu, T. et al. Cerebrospinal fluid GFAP is a predictive biomarker for conversion to dementia and Alzheimer’s disease-associated biomarkers alterations among de novo Parkinson’s disease patients: a prospective cohort study. J. Neuroinflamm. 20, 167 (2023).
Oeckl, P. et al. Glial fibrillary acidic protein in serum is increased in Alzheimer’s disease and correlates with cognitive impairment. J. Alzheimer’s Dis. 67, 481–488 (2019).
Yang, Z. & Wang, K. K. Glial fibrillary acidic protein: from intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 38, 364–374 (2015).
Wilson, H. et al. Imidazoline 2 binding sites reflecting astroglia pathology in Parkinson’s disease: an in vivo11C-BU99008 PET study. Brain 142, 3116–3128 (2019).
Herrera, M. L. et al. Early cognitive impairment behind nigrostriatal circuit neurotoxicity: are astrocytes involved? ASN Neuro 12, 1759091420925977 (2020).
Funahashi, S. & Andreau, J. M. Prefrontal cortex and neural mechanisms of executive function. J. Physiol. 107, 471–482 (2013).
Hallett, M. & Wu, T. The cerebellum in Parkinson’s disease. Brain 136, 696–709 (2013).
Huang, C. et al. Metabolic brain networks associated with cognitive function in Parkinson’s disease. NeuroImage 34, 714–723 (2007).
Zhan, Z. W. et al. Abnormal resting‐state functional connectivity in posterior cingulate cortex of Parkinson’s disease with mild cognitive impairment and dementia. CNS Neurosci. Ther. 24, 897–905 (2018).
Goldman, J. G., Williams-Gray, C., Barker, R. A., Duda, J. E. & Galvin, J. E. The spectrum of cognitive impairment in Lewy body diseases. Mov. Disord. 29, 608–621 (2014).
González-Redondo, R. et al. Grey matter hypometabolism and atrophy in Parkinson’s disease with cognitive impairment: a two-step process. Brain 137, 2356–2367 (2014).
Crucian, G. P. & Okun, M. S. Visual-spatial ability in Parkinson's disease. Front Biosci. 8, s992–s997 (2003).
Silbert, L. C. & Kaye, J. Neuroimaging and cognition in Parkinson’s disease dementia. Brain Pathol. 20, 646–653 (2010).
Pievani, M., de Haan, W., Wu, T., Seeley, W. W. & Frisoni, G. B. Functional network disruption in the degenerative dementias. Lancet Neurol. 10, 829–843 (2011).
Nelson, A. J. & Chen, R. Digit somatotopy within cortical areas of the postcentral gyrus in humans. Cereb. Cortex 18, 2341–2351 (2008).
Foo, H. et al. Progression of subcortical atrophy in mild Parkinson’s disease and its impact on cognition. Eur. J. Neurol. 24, 341–348 (2017).
Mavridis, I. N. & Pyrgelis, E. S. Nucleus accumbens atrophy in Parkinson’s disease (Mavridis’ atrophy): 10 years later. Am. J. Neurodegener. Dis. 11, 17–21 (2022).
Planche, V. et al. Anatomical predictors of cognitive decline after subthalamic stimulation in Parkinson’s disease. Brain Struct. Funct. 223, 3063–3072 (2018).
Picciotto, M. R., Higley, M. J. & Mineur, Y. S. Acetylcholine as a neuromodulator: cholinergic signaling shapes nervous system function and behavior. Neuron 76, 116–129 (2012).
Valkenborg, D., Rousseau, A. J., Geubbelmans, M. & Burzykowski, T. Support vector machines. Am. J. Orthod. Dentofac. Orthoped. 164, 754–757 (2023).
Postuma, R. B. et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 30, 1591–1601 (2015).
Litvan, I. et al. Diagnostic criteria for mild cognitive impairment in Parkinson’s disease: Movement Disorder Society Task Force guidelines. Mov. Disord. 27, 349–356 (2012).
Emre, M. et al. Clinical diagnostic criteria for dementia associated with Parkinson’s disease. Mov. Disord. 22, 1689–1707 (2007).
Bušková, J. et al. Validation of the REM sleep behavior disorder screening questionnaire in the Czech population. BMC Neurol. 19, 110 (2019).
Giladi, N. et al. Construction of freezing of gait questionnaire for patients with Parkinsonism. Parkinsonism Relat. Disord. 6, 165–170 (2000).
Tanaka, T. [[Fundamentals] 5. Python+scikit-learn for Machine Learning in Medical Imaging]. Nihon Hoshasen Gijutsu Gakkai Zasshi 79, 1189–1193 (2023).
Lundberg, S. M. et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2, 56–67 (2020).
Acknowledgements
This study was funded by the Applied Basic Research Foundation of Yunnan Province [Grant numbers 202301AS070045, 202101AY070001-115]; National Natural Science Foundation of China [Grant number 81960242]; The Major Science and Technology Special Project of Yunnan Province [Grant number 202102AA100069]; The Innovative Team of Yunnan Province [202305AS350019]; Shaanxi Provincial Natural Science Basic Research Program [Grant number 2024JC-YBQN-0822].
Author information
Authors and Affiliations
Contributions
Conceptualization: Z.Y.Y., W.F., P.A.L., and Y.X.L. Supervision: P.A.L. and Y.X.L. Data curation: N.P.P., L.K.L., Z.Y.F. and Z.L.F. Writing—original draft: Z.Y.Y. and W.F. Review and editing: L.B., R.H., X.Z., P.A.L., and Y.X.L. Investigation: Z.Y.Y., W.F., N.P.P., L.K.L., L.B. and R.H. All co-authors read and approved the document.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, Y., Wang, F., Ning, P. et al. Multimodal neuroimaging-based prediction of Parkinson’s disease with mild cognitive impairment using machine learning technique. npj Parkinsons Dis. 10, 218 (2024). https://doi.org/10.1038/s41531-024-00828-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41531-024-00828-6