Abstract
Identifying novel drugs that can interact with target proteins is a highly challenging, time-consuming, and costly task in drug discovery and development. Numerous machine learning-based models have recently been utilized to accelerate the drug discovery process. However, these existing methods are primarily uni-tasking, either designed to predict drug-target interaction (DTI) or generate new drugs. Through the lens of pharmacological research, these tasks are intrinsically interconnected and play a critical role in effective drug development. Therefore, the learning models must be utilized in such a manner to learn the structural properties of drug molecules, the conformational dynamics of proteins, and the bioactivity between drugs and targets. To this end, this paper develops a novel multitask learning framework that can predict drug-target binding affinities and simultaneously generate new target-aware drug variants, using common features for both tasks. In addition, we developed the FetterGrad algorithm to address the optimization challenges associated with multitask learning particularly those caused by gradient conflicts between distinct tasks. Comprehensive experiments on three real-world datasets demonstrate that the proposed model provides an effective mechanism for predicting drug-target binding affinities and generating novel drugs, thus greatly facilitating the drug discovery process.
Similar content being viewed by others
Introduction
The process of drug discovery revolves around finding, modifying, and designing new drugs that can interact with target proteins. This process requires extensive experimentation, which poses challenges like high cost and time consumption1,2,3. Therefore, many computational data-driven models have been used in this domain to reduce cost and time consumption. These models can be divided into two groups: predictive and generative models. Predictive models aim to predict drug-target interactions (DTI), while generative models are typically designed to generate new drugs. The DTI measures the interaction between the drugs and target proteins, which determines the therapeutic effects of drugs4 and has played a vital role in drug discovery by leading the way in finding cures for new diseases5. Moreover, DTIs have also gained significant attention in the context of drug repositioning, where existing drugs are used for new therapeutic indications, unlike traditional drug discovery, which involves developing new drugs from scratch6. Recently, Drug-Target binding Affinity (DTA) prediction is gaining more attention, as it can provide rich information on the strength of interactions between drugs and targets7. The DTA prediction task is mainly divided into two categories. The first one is binary classification approaches8,9,10,11, which determines whether there exists an interaction between the drug and the target. However, considering the nature of the problem, there is a need for extensive information. For instance, if an interaction between the drug and the target exists, then the strength information of the interaction is required. The second category is regression-based models, which predict the interaction strengths in terms of binding affinity values.
Many regression-based models were used to predict the binding affinities between drugs and targets previously. Among them, KronRLS12 model utilizes the Kronecker product of a drug-drug similarity matrix and a target-target similarity matrix to define similarities between drug-target pairs. However, this model can only capture the linear dependencies. The SimBoost13 approach uses a nonlinear method based on gradient machines, which relies on features derived from drugs, targets, and drug-target pairs. The KronRLS and SimBoost may overlook the latent features present in the original drug and target. Besides machine learning models, deep learning-based models are also applied to this task. DeepDTA14 is a representative deep model, which utilizes a 1D Convolutional Neural Network (CNN) architecture to process both the drug SMILES and protein sequence. However, the model lags in extracting the relevant features via 1D CNN blocks for both the drugs and targets. On the other hand, WideDTA15 takes advantage of incorporating text-based information into the architecture. By integrating text-based information, WideDTA has demonstrated an improved performance over DeepDTA. However, both of these models still fail to capture drug molecule structural information. To overcome the issues, GraphDTA16 represents the drugs using a graph representation. By taking advantage of the graph’s representation of drugs, the model has witnessed a slight improvement in the results over the previous ones. However, this model has the limitation of using a limited set of atom features for the drugs. Moreover, CoVAE17 is proposed to predict the DTA and generate new drugs by taking advantage of Co-Variational autoencoder for both the drugs and targets. However, this model uses separate feature spaces for each task. Moreover, for the DTA prediction task, the CoVAE utilized the latent features extracted after variational layers, which sometimes miss the actual representation of drugs and targets. On the other hand, several generative models such as generative adversarial networks18,19, autoregressive models20, variational autoencoders17, and reinforcement learning21 have also been used in this domain. These models can generate a wide array of drug combinations without incorporating knowledge of the binding dynamics between the drugs and target receptors.
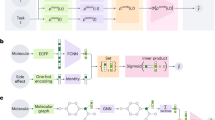
In this work, we propose a multitask learning (MTL) framework DeepDTAGen, which performs both tasks (predicting DTA and generating novel drugs) simultaneously by using the common features (as the knowledge of ligand-receptor interaction) space for both functions as reflected in Fig. 1. Minimizing the loss in the DTA prediction task ensures the learning for DTI-specific features in the latent space while utilizing these features in the drug generation task ensures the generation of target-aware drugs, thereby significantly increasing their potential for clinical success. However, MTL models are often prone to optimization challenges such as conflicting gradients22. To address this issue in DeepDTAGen, we developed the Fetter Gradients (FetterGard) algorithm to mitigate gradient conflicts. Primarily, unlike the existing methods, the DeepDTAGen has the following foundational novelties: (i) The proposed model uses a shared feature space and performs these tasks in a unified model. (ii) This study develops the FetterGrad algorithm, which keeps the gradients of both tasks aligned while learning from a shared feature space. It mitigates the gradient conflicts and biased learning by minimizing the Euclidean distance (ED) between task gradients. (iii) The DeepDTAGen offers two objective functions: it predicts drug-target affinity values while simultaneously generating target-aware drugs conditioned by the input interactions. The results through comprehensive experimentation demonstrate that DeepDTAGen not only accurately predicts binding affinity between drugs and targets but can also successfully generate target-aware drugs. In particular, we show the robustness of the DeepDTAGen in the DTA prediction through (i) drug selectivity, (ii) Quantitative Structure-Activity Relationships analysis, and (iii) Cold-start tests. Similarly, for the generative task, we perform (i) chemical drugability analysis, (ii) target-aware, and (iii) polypharmacological analysis of the generated drugs. We believe that DeepDTAGen provides a flexible strategy for the drug discovery process by predicting drug-target affinity and novel target-oriented drug generation.
Results
This section discusses the performance of DeepDTAGen on KIBA, Davis, and BindingDB datasets with a comparison to the state-of-the-art methods. For the Affinity prediction task, the Mean Squared Error (MSE), Concordance Index (CI), R squared r2m, and Area under precision-recall curve (AUPR) are evaluation metrics used to measure the performance of the proposed model. Each metric is discussed in detail in Supplementary Discussion, and the experimental setup is listed in Supplementary Table 3. Further, to evaluate the generative performance of the proposed model, we assessed the Validity, Novelty, Uniqueness, and their binding ability to their targets. The Validity measures the proportion of chemically valid molecules among all generated ones. Novelty calculates the proportion of valid molecules that are not present in the (Modified Target SMILES, MTS) target SMILES of both the training and testing sets. Uniqueness provides the proportion of unique molecules among the generated chemically valid ones. Moreover, we also performed chemical analyses on generated drugs with three chemical properties (Solubility, Drug-likeness, and Synthesizability), and structural analysis (including the counts of atom types, bond types, and ring types). Supplementary Discussion provides detailed explanations for each chemical property. We generated the SMILES with two distinct strategies: On SMILES and by Stochastic methods. In the first method, we generated SMILES by feeding the condition and original SMILES to the transformer decoder. However, in the Stochastic method, the model produces stochastic elements instead of the original SMILES, while the rest of the input conditions remain the same. The first method allows the researchers to consider the broader spectrum of potential drug candidates. However, the second method provides the solution to generate SMILES for specific target proteins. The first seven subsections of the results discuss the performance of the binding affinity task, while the remaining subsections discuss the performance of the generative task.
Predictive performance (binding affinity)
In Table 1, we presented the results comparison between DeepDTAGen and the existing DTA prediction models on three benchmark datasets, while Fig. 2 shows the predicted affinities of all the test sets. On the KIBA test set, DeepDTAGen achieved results of 0.146, 0.897, and 0.765 in terms of MSE, CI, and \({r}_{m}^{2}\), respectively. Similarly, on the Davis test set, the model attained the MSE of 0.214, CI of 0.890, and \({r}_{m}^{2}\) of 0.705. Whereas on the BindingDB test set, the proposed model achieved 0.458, 0.876, and 0.760 of MSE, CI, and r2m, respectively. The DeepDTAGen model outperforms traditional machine learning models (KronRLS and SimBoost) on the KIBA dataset by gaining the improvement of 7.3% in CI and 21.6% in \({r}_{m}^{2}\), and the MSE is reduced by 34.2%. However, compared to the deep learning models, especially the second-best model (i.e., GraphDTA), DeepDTAGen attained an improvement of 0.67% in CI and 11.35% in \({r}_{m}^{2}\) while reducing MSE by 0.68%. The proposed model also compromised the CI of 2.3% compared to the GDilatedDTA23. While comparing the results obtained on the Davis dataset, the performance of the DeepDTAGen surpasses the traditional machine learning models. Specifically, we witness the improvement of 2.0% in CI and 9.4% in r2m, while the mean squared error is reduced by 24.1%. Moreover, when compared with the second-best deep learning model SSM-DTA, the DeepDTAGen has the edge of 2.4% improvement in terms of \({r}_{m}^{2}\), 2.2% drop in MSE. Finally, on the BindingDB dataset, the proposed model gained an improvement of 0.9% in terms of CI and 4.1% in \({r}_{m}^{2}\) while a 5.1% decrease in terms of MSE compared to the GDilatedDTA.
Throughout all comparisons, the proposed model consistently suppresses the previous models like DeepDTA, CoVAE-DTA, WideDTA, AttentionDTA, DeepCDA, GDilatedDTA, and ELECTRA-DTA. This can be primarily attributed to these models’ reliance on the string representation of molecules, which tends to lack in capturing the structural information of the molecules. In contrast, models like GraphDTA, DoubleSG-DTA, and GDilatedDTA have the advantage of using drugs as graph representation. However, these models utilize a few node (atom) features of the drugs. Considering this issue, we have incorporated additional DTI-centric node features in the DeepDTAGen, resulting in a more comprehensive and informative representation of the drug molecules. In addition, we use an NLP-based model (Gated-Convolution neural network, Gated-CNN) to extract features from protein sequences, which allows the model to learn the key parts while discarding irrelevant features. These enhancements to our model led to significant improvements in the results.
Model tests on a drug selectively
In this phase, we have conducted drug selection tests. The following criteria have been set for considering a drug in the test: (1) The drug has high variability in binding affinity value with targets. (2) The drug that interacts with the targets with more or less same binding affinity. Considering the first criteria in the KIBA dataset, we have selected the drug 3-[3-[2-[4-(4-ethylpiperazin-1-yl)anilino]pyrimidin-4-yl]imidazo[1,2-a]pyridin-2-yl]-N-(2-fluorophenyl)benzamide which has a high degree of variation among its affinity values to the target proteins. The chosen drug has the highest affinity with the Q9HAZ1 and the lowest affinity with P43405 proteins, while medium binding affinity with the Q9H2X6 protein. The DeepDTAGen has also accurately predicted the respective levels of affinity for the selected drug with these target proteins (Supplementary Table 6). Similarly, in the Davis dataset, we chose (1-N’-[3-fluoro-4-[6- methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxyphenyl]-1-N-(4-fluorophenyl)cyclopropane-1,1 dicarboxamide) drug. The selected drug has a higher affinity with Kit_Human protein, medium affinity with target protein MTIK, and lower affinity with PIP5K1C. The DeepDTAGen has successfully predicted the same affinity levels with these specific interactions (Supplementary Table 7). Moreover, in the BindingDB dataset, we chose 2-[(3,5-dimethoxyphenyl)methyl]-5-(4-fluoro-2-methylphenyl)-7-[(2-imino-1,3-thiazolidin-3-yl)methyl]-3,4-dihydroisoquinolin-1-one drug. The selected drug binds to the P61964 protein by higher affinity than its other interactions, while the DeepDTAGen maintained its level by predicting the same level of affinity to the specific protein in Supplementary Table 8.
Taking the second criterion into account, we have selected the Kenpaullone drug from the KIBA dataset, which has similar affinities with the PAK4_HUMAN, TYK2, and MK01_HUMAN proteins. DeepDTAGen has successfully maintained the position of these interactions by predicting the more or less same affinity values with the variance of 0.04, 0.01, and 0.07 for PAK4_HUMAN, TYK2_HUMAN, and MK01_HUMAN proteins, respectively (Supplementary Table 9).
Furthermore, in the Davis dataset, we selected the (N-[4-(3-chloro-4-fluoroanilino)-7-(oxolan-3-yloxy)quinazolin-6-yl]-4-(dimethylamino)but-2-enamide), drug with a similar binding affinity with TAK1, RAF1, and RPS6KA5 KinDom.1-N-terminal proteins (Supplementary Table 10). The DeepDTAGen has proven its validity by maintaining its ranks and predicting the same affinity values with a variance of 0.08, 0.1, and 0.1. Moreover, we select 2R-1-[4-[(4-fluoro-2-methyl-1H-indol-5-yl)oxy]-5-methylpyrrolo[2,1-f][1,2,4]triazin-6-yl]oxypropan-2-ol from the BindingDB that have similar affinities with the Q9HBH9, P04626, and O96013proteins (Supplementary Table 11). The proposed model has successfully maintained the position of these interactions by predicting the more or less same affinity values with a variation of 0.06. In addition, a graphical illustration of various selective drug tests is presented in Supplementary Figs. 14–16.
Randomization tests for model validation
Here, we evaluated the validity of the proposed model using four randomization tests. These tests aimed to investigate whether the observed results reflect the true biological correlation between the drugs and targets or if they are obtained through random chance. These tests include y-randomization, Drug randomization, Protein randomization, and Protein Descriptor randomization across the KIBA, Davis, and BindingDB datasets. In the y-randomization test, we randomly shuffled the affinity values associated with the interactions, while keeping the interactions themselves unchanged. In the same way, the drugs were randomly shuffled, while the target proteins and affinity values remained unchanged in the Drug Randomization test. In the Protein Randomization test, we permuted the target proteins only, leaving the rest of the data unchanged. Finally, in the Protein Descriptor Randomization test, we replaced the target proteins with random strings. Next, we trained the proposed model on these permuted datasets, and their results are evaluated under our standard evaluation metrics. Supplementary Table 12 lists the results of each permuted dataset along with the comparison with the standard datasets, while Supplementary Fig. 17 shows the scatter visualization of predicted affinities versus the original affinities of each randomization test on KIBA, Davis, and BindingDB datasets. In Supplementary Table 12, it can be seen that in the y-randomization test (where the affinity values were shuffled), the model performance declined to near-random levels across all three datasets. The CI dropped to around 0.5 for all datasets, while the MSE increased significantly, and \({r}_{m}^{2}\) and AUPR dropped to near 0 across all the permuted datasets. Similarly, in the Drug Randomization test, the performance is noticeably declined with higher MSE and lower CI, \({r}_{m}^{2}\), and AUPR values. In the same way, in the Protein Randomization experiment, the AUPR dropped to 0.0 on the BindingDB dataset, while the MSE increased throughout all the datasets. Moreover, in the Protein Descriptor Randomization test, MSE increased to 0.572 on the KIBA permuted dataset, while on Davis and BindingDB, MSE rose to 0.458 to 1.586, respectively. The CI and \({r}_{m}^{2}\) were also decreased to 0.655, 0.670, and 0.680 for KIBA, Davis, and BindingDB, respectively. Notably, we observed a significant decrease in AUPR to 0.111 on the permuted BindingDB dataset. Overall, the results of these randomization tests suggest that the proposed model does not rely on spurious correlations or chance. Instead, it effectively learns the drug-target relationships, which supports the validity of our model’s hypothesis.
Cold-start affinity test
To evaluate the proposed model’s performance in cold-start scenarios, we utilized two splitting methods: drug-wise and protein-wise splitting on the KIBA, Davis, and BindingDB datasets. In the Drugs-wise splitting, we identified the unique drugs in each dataset and divided them into training and testing sets with ratios of 80:20, respectively. Similarly, for the Proteins-wise split, we applied the same protocol, with the division based on unique target proteins rather than drugs. In both settings, the division of the training and test sets was fully randomized. Supplementary Fig. 18 shows the predicted affinities by the proposed model compared to the actual affinities, using drug-wise and protein-wise splitting methods for each dataset. Moreover, Supplementary Table 13 presents the results of the proposed model with the comparison to the previous deep learning-based DTA models under these two splitting methods using the standard evaluation criterion. It can be seen from the table that in the Drugs-wise splitting method, on the KIBA dataset, the Affinity2Vec model performed better. However, on both the Davis and BindingDB datasets, our proposed model demonstrated the best performance across all models in terms of MSE and \({r}_{m}^{2}\). On the other hand, in the Proteins-wise split, the DeepDTA model performed better on the Davis dataset. However, on the KIBA and BindingDB datasets, our model achieved better performance, with the least MSE and higher \({r}_{m}^{2}\) and AUPR. These results suggest that our model is more robust than the previous models towards unseen data.
Ablation study and hyperparameter tuning
To validate the effectiveness of DeepDTAGen, we also conducted a series of five ablation tests presented in Supplementary Table 4. In the first experiment, the After the Mean and Log Variance Operation (AMVO) features for drugs were used. In the second experiment, limited atomic features of drug graphs were utilized, similar to the GraphDTA16. In the third test, the Gated-CNN in the Gated-CNN Module was replaced with a 1D CNN. The fourth experiment was conducted by replacing the Graph Convolutional layers with the 1D CNN layers in the Encoder module. In the fifth test, the GCLs and Gated-CNN layers in the Encoder and Gated-CNN modules were replaced with 1D CNN layers. Experiment 1 showed worse performance than the rest of the ablation tests. As in the test, latent features representation was obtained AMVO. The results achieved through the 5th experiment were also inferior, mainly due to the replacements of GCLs and Gated-CNN layers with 1D CNN in Encoder and Gated-GNN modules, which restricted the model from learning the structural features of the input. The fourth experiment showed slightly better performance over Model 5 due to the induction of Gated-CNN in the Gated-CNN module. The results of the third test are superior to the aforementioned tests, due to the utilization of the graph representation for the molecules, endorsed by graph convolutional layers, and taking advantage of additional node features to enhance the representation of the drug nodes. Finally, the best results were achieved in all the ablation tests in the second experiment. The ablution study shows that combining different configurations, like representing drugs through graphs, using Gated-CNN on the proteins side, and considering additional features of drugs, significantly improves prediction performance. Further, in Supplementary Discussion, we presented the results obtained by our model with different network hyperparameter settings.
Validity, Novelty, and Uniqueness of generated drugs
Here, we compared the generative performance of the proposed model to previous models in terms of Validity, Novelty, and Uniqueness. For a fair comparison, we retrained previous models in our environment on the KIBA and BindingDB datasets, as these models were previously trained on other datasets. Due to the minimum amount of unique drugs available in the Davis dataset, including the proposed model, none of the existing models generated novel drugs on the Davis dataset. The comparative results are summarized in Table 2. Among them, CoVAE17, ORGAN19, SMILES LSTM24, and Syntalinker25 showed the lowest performance on both KIBA and BindingDB datasets, while the performance of PGMG20 is comparable to our proposed model. On the KIBA dataset, the proposed model outperforms the PGMG with an improved validity ratio of 3% in validity ratio. However, the novelty and uniqueness rate of the proposed model are comparatively lower than PGMG. On the other hand, on the BindingDB dataset, the proposed model surpasses the PGMG with improvements of 4.9% and 10% in terms of validity and novelty ratios, however, we noticed a significant drop in the uniqueness ratio. These results suggest that the proposed model remained successful in generating valid and novel drugs. However, the lower uniqueness ratio can be attributed to the polypharmacological nature of training data, where a drug interacts with multiple target proteins, and proteins interact with many drugs. Therefore, the input consists of drug-protein interaction pairs, and the target output for the model is often the same drug across multiple interactions. On the other hand, second-best model PGMG utilizes randomized SMILES as target SMILES, which are completely different from input SMILES. Such a method can result in an improved uniqueness ratio but compromises the learning of biological relevance between drugs and targets.
Interaction-based drugs generation (on SMILES)
In this method, drugs were generated using drugs and their target proteins as input conditions. After the generation process, we predicted the binding affinities of the generated drugs with their target proteins, which were used as conditions for generating the drugs. For a fair analysis, we used AutoDock VINA26 as a proxy for affinity predictions. Figure 3 shows the distribution of binding affinities between generated drugs and their seed targets, calculated in Kilocalories per mole (kcal/mol), where the lower value indicates the stronger binding. Figure 3A shows the affinity distribution for KIBA-generated drugs, while Fig. 3B represents the affinity distribution for BindingDB. The figure demonstrates that both distributions exhibit satisfactory affinities to their respective seed targets, with a very minimal number of interactions having weak bindings. Moreover, Fig. 4, displays four drugs from the generated set of KIBA and BindingDB, that have suitable drug-like properties. Furthermore, In Fig. 5, we visualized the docking sites of these drugs to their corresponding targets (seed targets) in comparison to the original drugs. As shown in the figures, both the generated and the original drugs exhibit the same binding sites as per Uniprot27. These results suggest that the proposed model successfully generates target-aware drugs.
The first column in the figure represents the trained model. The second column shows the PubChem ID for the drugs and the UniProt ID for the targets (both the drugs and targets are used as seeds to generate new SMILES). The third column lists the chemical structure of the seed SMILES. The fourth column shows the chemical structure of the generated SMILES. The fifth column shows the Tanimoto Similarity (TS) between the generated and seed drugs. The sixth column displays the chemical properties of the generated drugs. The last column shows the Docking Scores (DS) for the seed target with the seed drug and the generated drug, where the seed value represents the DS between the seed drug and the seed target, while the generated value represents the DS between the generated drug and the seed target.
A–D (labeled “KIBA” in the figure) represent the binding sites for the KIBA-generated and their seed drugs, corresponding to rows 1–4 of the table. A–D, which are labeled as “BindingDB” in the figure, represent the binding sites for the BindingDB-generated and seed drugs, corresponding to rows 5–8 of the table. The red folds in the figure are the binding sites for the respective targets as per the uniport database.
Evaluation of the chemical properties of the generated drugs
In this section, we performed the chemical analysis on the generated drugs. Figure 6 depicts the chemical similarity between the generated and test set drugs from the KIBA dataset, while Supplementary Fig. 19 shows the chemical similarity between the BindingDB test set and generated drugs. It can be seen in Fig. 6 that the generated drugs (On the SMILES method) have the acceptable average distribution of QED, LogP, and SAS of 0.519, 3.391, and 2.721, respectively, on the KIBA dataset. The generated drugs on the BindingDB have the QED, LogP, and SAS of 0.325, 3.427, and 3.271, respectively. On the other hand, through the Stochastic method, consistent results are observed on both datasets. Specifically, in one KIBA set, the average distribution remained 0.502, 3.756, and 2.776. Similarly on the BindingDB set, these averages are 0.592, 2.391, and 2.478 of QED, LogP, and SAS, respectively. Furthermore, we evaluated the generated drugs against Lipinski’s Rule of Five, a widely used guideline in drug discovery for assessing physicochemical properties essential for oral bioavailability28. These rules include drugs having five or fewer hydrogen bond donors, a molecular weight of less than 500 Da, a partition coefficient (logP) of less than five, and ten or fewer hydrogen bond acceptors. As can be seen in Supplementary Fig. 20 that most of the generated molecules satisfy Lipinski’s Rule of Five.
A The QED, LogP, and SAS properties distributions in the original KIBA test set. B The same QED, LogP, and SAS properties distribution in the generated molecules by on SMILES synthesis method, C the distribution of generated molecules using the stochastic method. In each panel, the notation μ represents the mean of that distribution.
Additionally, we analyzed the structural features between the generated and original drugs. These structural features include atomic types, bond types, and ring types. Supplementary Fig. 21 shows the structural features of the generated drugs and the original from the KIBA dataset, while Supplementary Fig. 22 represents the BindingDB dataset. The distribution of the structural features in the generated drugs by the On SMILES method is comparable to or even exceeds those distributions in the test sets. However, the distribution of structural features in generated drugs on the Stochastic method is lower than the distribution in the test sets. It could potentially be attributed to the provision of SMILES strings as input instead of stochastic elements, where the model remains more focused on the structurally coherent molecules based on the provided condition (SMILES strings).
Drug generation on the stochastic method
In this section, we evaluated the DeepDTAGen in drugs generation through the stochastic method. As we mentioned earlier, the stochastic method considers only the target sequence as input, while the SMILES are replaced with the stochastic elements produced by the model. The method allows the researchers to generate drugs for different target proteins. As a test case, here we consider the Epidermal Growth Factor Receptor (EGFR) PDBID: P00533, which is highly responsible for cell lung cancer29. We then proceed to generate drugs using the EGFR sequence as input to both KIBA and BindingDB pre-trained models. Once the drugs were generated, we then assessed their chemical properties by considering parameters such as QED, LogP, and SAS. Further, we used AutoDock VINA26 to evaluate their interaction with the EGFR target protein. Figure 7 displays the generated SMILES, the 2D and 3D structures of drugs, and the pocket area of the EGFR protein, along with their QED, LogP, and SAS scores. It can be seen in Fig. 7 that the generated drugs through both pre-trained models are successfully attached to the EGFR protein. Moreover, the red highlighted folds in both panels of the figure represent the attached amino acids to the generated drugs, and these folds are the binding residues according to the UniProt library27. The results indicate that the proposed model can generate target-aware drugs with favorable chemical properties.
Polypharmacological druggability evaluation of generated drugs
In drug discovery research, one of the emerging paradigms is polypharmacology30,31,32, where a single drug responds to multiple targets of a specific disease, or a single drug acts on multiple targets for multiple disease pathways33. Therefore, understanding the polypharmacological effects of the molecules is crucial for investigating their off-target effects and drug selectivity, specifically in the context of multi-target drug design and drug repurposing. In this regard, we evaluated the polypharmacological effects of some generated drugs. Accordingly, we selected five drugs from the generated molecules of KIBA and BindingDB sets, having suitable drug-like properties, and their seed drugs interacted with at least three other target proteins. We then predicted the docking scores of these generated drugs with their seed proteins as well as with three other proteins that were active against their respective seed drugs using AutoDock VINA. Figure 8 shows a comparison of docking scores for KIBA, while Supplementary Table 14 provides a comparison of docking scores for BindingDB. In both comparisons, the affinities of generated drugs for the three other target proteins are somewhat lower than their seed proteins. Yet, the generated drugs maintained their polypharmacological effects for the targets that were active against the seed drugs. These results suggest that, despite being generated for specific interactions, these drugs still retained their polypharmacological effects, which shows their ability for practical applications in drug discovery.
The first column in the figure shows the PubChem ID for the drugs and the UniProt ID for the targets (both the drugs and targets are used as seeds to generate new SMILES). The second column lists the chemical structure of the seed SMILES. The third column shows the chemical structure of the generated SMILES. The fourth column shows the Tanimoto Similarity (TS) between the generated and seed drugs. The fifth column displays the chemical properties of the generated drugs. The sixth column shows the other active targets against the seed drug. The seventh column represents the Docking Score (DS) between the three corresponding targets and the generated drug. The last column represents the Docking Score (DS) of the seed target with the seed drug and the generated drug.
DTI-driven guidance on target-aware drug generation
Here, we experimented to validate that the DTI task guides latent space to extract target-aware features. In this regard, we trained the model by eliminating the prediction task and focusing only on the generative task. The drug encoder, latent space, and the decoder from the proposed model were used in this setting. Once trained, we generated new drugs SMILES on both datasets. For each generated drug, we predicted its docking score (using AutoDock VINA with the default parameters for a fair comparison) with the target proteins that were active against the seed SMILES (the original SMILES used as input to generate the new SMILES) in the dataset. In the same way, the docking scores were predicted between the generated SMILES by DeepDTAGen and their seed target proteins. For certain interactions, the simple encoder-decoder model did not generate valid drugs. Therefore, to ensure a fair comparison, we also excluded the drugs generated using those interactions as seeds by the DeepDTAGen. Supplementary Fig. 23 shows the comparison of docking score distributions for drug generation between the multitask DeepDTAGen model and the single-task framework. It can be seen in the figure that the distribution of the docking scores by DeepDTAGen generated drugs is comparatively higher than the single-task learning framework, which validates our model hypothesis.
Discussion
In this study, we proposed a novel multitask learning framework that offers two objective functions: predicting DTA and generating novel drugs. The main objective of performing these tasks within the unified model is to enable the model to learn the biological relationships between drugs and targets. In the proposed model, both tasks are performed from the shared feature space, suggesting that the learned features are highly correlated with both drugs and targets. Furthermore, we introduced the FetterGrad algorithm, which mitigates biased learning and resolves gradient-conflicting issues between the DTA prediction and drug generation tasks. To validate our model hypothesis, we perform extensive experiments in DTA prediction and drug generation tasks.
In the DTA prediction task, we show that the affinity predictions by the proposed model are aligned with biologically relevant behavior between drugs and targets by performing drug selectivity analysis where two drugs were selected from each dataset based on their affinity profiles: one with high variations in affinity with different targets and another with consistent affinity levels with its targets. Our model successfully predicted accurate affinities for both drugs, which highlights the model’s ability to learn the biological patterns of DTI. Similarly, in another test, we performed four randomization experiments, where we corrupted the associations between drugs and targets to validate the model’s ability to learn genuine relationships between them. The comparison of results obtained from the standard and the permuted datasets demonstrated that the model successfully learned the actual relationships between drugs and targets.
On the other hand, in the generative task, the proposed model remained successful in generating target-aware drugs. To evaluate whether the DTA task effectively guides the network in generating these target-aware drugs. In this regard, we conducted an experiment in which the model was trained with and without the DTA task. The results showed that the drugs generated with the DTA task exhibited stronger affinity to their seed targets compared to those generated without the DTA task. Although the DeepDTAGen performed well on both tasks, it still has the limitation of lacking the support of chemical properties such as QED, LogP, and SAS as conditions. Secondly, the proposed model ignores the stereochemistry dynamics of the input molecules, while stereochemistry plays an important role in drug development and discovery. Therefore, the consideration of these properties, along with an appropriate guidance mechanism and embedding the stereochemistry information, will be a valuable extension. Furthermore, the model can also be extended by incorporating non-interacting data during training, which could enhance drug generation with higher selectivity.
Methods
Datasets
We used the KIBA34, Davis35, and BindingDB36 datasets to evaluate our model performance. These datasets are commonly considered as benchmarks for drug-target affinity prediction, which are also used by other state-of-the-art models like DeepDTA, GraphDTA, and CoVAE. In all the datasets, the drug SMILES strings were extracted from the PubChem database37 using the PubChem CIDs. The target protein sequences were extracted from the UniProt database38 based on their gene names. The Davis dataset consists of 68 drugs and 442 targets. Each drug is paired with the target protein by affinity value measured as Kd (kinase dissociation constant). Initially, the affinity values have a large variability, which may affect the model performance. Therefore, we transformed the Kd values into the range of 5.0–10.8, using the negative logarithm (pKd) shown in Eq. (1).
The KIBA dataset contains 2111 drugs and 229 targets, with a total of 118254 interactions; the range of affinity values is 0.0–17.2. In the BindingDB dataset, the IC50 scores are considered as the affinity between drugs and targets. In total, this dataset consists of 18,044 unique drugs and 1620 unique proteins, with the total of 56,525 interactions. More details about the datasets are listed in Supplementary Table 1. Moreover, we followed the GraphDTA method by dividing all the datasets into training and testing sets. Each dataset consists of six folds, with one fold considered as the test set, and the remaining five are used as the training set. By dividing the data, we have kept the testing set completely separate from the training set. The distributions of affinity values for the KIBA, Davis, and BindingDB datasets are shown in Supplementary Figs. 1–3.
Feature representation
Protein representation
In all the datasets, each protein sequence is encoded as a series of ASCII characters, representing the building blocks of each amino acid. We applied the label encoding method by assigning different numerical values to the ASCII characters based on their alphabetical symbols. To regularize the length of each sequence uniformly, we applied padding and trimming methods. The standard length of 1000 is set for each sequence. If the sequence is shorter than the standard length, it is padded with zeros. Similarly, if a sequence is longer than the standard length, the characters exceeding the limit are trimmed, as illustrated in Supplementary Fig. 4. Furthermore, we have extracted a 128 × 1000 dimensional matrix against each numerical sequence using the word embedding method, which is the input to our model, further detail is available in the Supplementary Methods.
Drugs representation
We used two representations of the drugs, SMILES; Graph representations of drugs as the input of the Graph-Encoder Module, and MTS strings as the target objective of the Transformer-Decoder Module. In both graphs and MTS strings representation, we consider only SMILES with lengths equal to or less than 138 for training. SMILES exceeding the specified length are discarded along with their corresponding interactions. Moreover, for the graph representation, we have converted the SMILES strings from canonical to the isomeric form, similar to the GraphDTA using the standard chemical RDKit library39. Furthermore, we converted the isomeric drug strings to nodes and edges. Since our goal was to represent drugs with comprehensive information in graph representation, therefore we have extracted a comprehensive set of features of the nodes (atoms) using the RDKit library39, which includes ring membership, hybridization, formal charge, the implicit value of the atom, adjacent hydrogens, adjacent atoms, aromatic structure, and atom symbol. Each of the atom features and their importance in the context of DTI is discussed in detail in Supplementary Methods. Moreover, Supplementary Table 2 lists the features with the corresponding encoding method and their dimension, while the conversion of SMILES strings to graph representation and the features encoding method are illustrated in Supplementary Figs. 5 and 6. For the MTS, we performed modifications to the Target SMILES strings based on the QED score. SMILES with less QED score are injected with a set of spatially chemical features, which are essential for DTI. In the modification phase, we ensured that no SMILES remained invalid, no SMILES had disconnected atoms, and their scaffold remains valid. More details about the spatially chemical features are available in the Supplementary Methods.
Model foundation
As depicted in Fig. 1, the proposed framework has four modules: a Gated-CNN module for extracting features from protein sequences, a Graph-Encoder Module for capturing structural features of drugs, a Fully-Connected Module for affinity prediction, and a Transformer-Decoder module for generating drug molecules. We first converted the drug SMILES strings into graph representations with comprehensive node features (atomic-level features essential for drug-target binding). These drug graphs are then processed by the Graph-Encoder module while the protein sequences are fed to the Gated-CNN module. The features extracted by the Graph-encoder module are categorized into two feature sets: Prior to the Mean and Log Variance Operation (PMVO) and AMVO. The PMVO features are concatenated with the features of target proteins (extracted by the GCNN-block) and passed to the Fully-Connected Module for affinity prediction. Similarly, the AMVO features are fed to the multi-head cross-attention block of the Transformer-Decoder along with the corresponding affinity values and protein vectors (extracted by the GCNN-block) as Key (K) and Value (V). Moreover, the Transformer-Decoder Module receives the embeddings of MTS after positional encoding and embedding operation as input. Where Query (Q) is derived from these MTS embeddings, and K, V is obtained from the combination of AMVO, Affinity value, and the output of the Gated-CNN module for performing cross attention. This cross-attention mechanism ensures that the decoder effectively focuses on the shared features of the drug and protein data, enabling the generation of novel and target-specific drug molecules. Finally, the loss function of the Fully-Connected Module is defined based on the error between the actual affinity values and the predicted ones. Similarly, the loss of the Transformer-Decoder Module is calculated based on the error between the original MTS and the reconstructed SMILES. We formulated the drug-target affinity prediction task and novel drug generation in the following subsections and illustrated them graphically in Supplementary Fig. 8, while the model specification is presented in Supplementary Methods.
Gated-CNN module
To predict the protein-ligand binding affinity, we utilized the Gated-CNN, which adds a gate after each convolutional layer. The gating mechanism allows the model to selectively update and forget information, which enables the model to learn long-term dependencies in sequences effectively. The Gated-CNN receives the word embedding matrix and forwards it to the gated convolutional layer, where the matrix is split into two parts40, unlike a simple 1D CNN. One part is the convolution value (CV-Unit), and the second is the gated value (GV-Unit). The GV-Unit reacts as a gate to control the CV-Unit output by passing through the sigmoid function, transforming the GV-Unit value from 0 to 1 as output. A value near 1 shows that the information is relevant or more important, whereas a value near 0 indicates that the information is irrelevant or less important. The final output (ZTargets) is the element-wise product of CV-Unit and Sigmoid GV-Unit. Mathematically, it can be represented as:
In both equations, XTargets is the input parts for CV-Unit and GV-Units, W is the concern weights matrices, and c are biases. Since the GV-Unit is passed through the Sigmoid activation function as:
Here, the σ is the sigmoid activation function. The final output is the element-wise product of CV-Unit and Sigmoid(GV-Unit), which can be defined as follows:
Graph-Encoder and Transformer-Decoder Modules
The Encoder and Decoder are the backbone of our proposed model, responsible for synthesizing drugs and directing the Fully-Connected Module for binding affinity prediction. The Whole operation can be formulated as follows: Let XDrug is the graph representation of a drug, and C = {ZTarget, Y} is the condition vector. Then the Graph-encoder \(q({{\bf{Z}}}| {X}_{\,{\mbox{drug}}}^{i},{A}_{{\mbox{drug}}\,}^{i})\) encodes the node feature vector \({X}_{\,{\mbox{drug}}\,}^{i}\) and the corresponding adjacency matrix \({A}_{\,{\mbox{drug}}\,}^{i}\) into the latent vector Z. Where the condition vector C is added to the latent vector like Zcondition = Concat(Z, C). Further, the conditional latent vector Zcondition is forwarded to the Multi-Head cross-attention component of the Transformer-Decoder Module. Similarly, the Transformer-Decoder Module p(DrugSMILES∣ZCondition,MTS) takes latent vector ZCondition and SMILES string MTS as input and generates target SMILES.
Building on the above initial explanation, the Graph-encoder \(q({{\bf{Z}}}| {X}_{\,{\mbox{drug}}}^{i},{A}_{{\mbox{drug}}\,}^{i})\) takes the node feature vector \({X}_{drug}^{i}\) and the adjacency matrix \({A}_{drug}^{i}\) of a drug as input and transforms it to a low-dimensional representation through a couple of GCN layers using the following expression:
where D is the degree matrix, which contains information about the number of edges connected to each node, W0 is the weight matrix, and ReLU is the activation function. Here, the feature vector Xi serves a dual purpose in both affinity prediction and drug generation tasks. In the context of affinity prediction, the exact Xi feature vector is utilized. However, for drug generation, these feature vector is passed through multiple transformation stages. Like, we transformed the graph representations Xi to sequences \({S}_{\,{\mbox{drug}}\,}^{i}\) to ensure their compatibility with the transformer decoder. Following the PGMG20 segment encoding is added to the sequence like \({{{\bf{S}}}}_{\,{\mbox{drug}}}^{i}={S}_{{\mbox{drug}}\,}^{i}+SE\) where SE is the segment encoding.
Furthermore, the mean (μ) and log variance (σ) operations in the subsequent variational layers are applied to distribute the input Sdrug; into a range of 0 to 1 multivariate Gaussian distribution. The latent vector representation Z is derived by sampling from the distribution, reflected in Eq. (7). Where ϵ is a random noise vector produced by a normal distribution N(0, I) with the mean 0 and variance 1. It is used as a reparameterization trick, which involves adding the element-wise product to the covariance matrix \(\sigma ({{{\bf{S}}}}_{\,{\mbox{drug}}\,}^{i})\). Essentially, the reparameterization trick enables the backpropagation of gradients through stochastic sampling41, which is illustrated in Supplementary Fig. 9. Finally, at this stage, the condition C = {ZTarget, Y} is added with a latent vector as follows:
Ultimately, the Graph-Encoder Module returns two outputs Zcondition (AMVO) and X (PMVO), which are fed to the Transformer-Decoder and Fully-Connected Modules, respectively.
Whereas, the transformer decoder uses latent variables Zcondition and MTS as input. However, the MTS is first passed through several steps, including tokenization, embedding, and positional encoding. Subsequently, the resultant vector is projected into Query (Q), Key (K), and Value (V) vectors with dimensions of dmodel. The projected vectors are passed through the Masked Multi-Head Attention (MMHA) sublayer of the transformer decoder. This sublayer utilizes attention mechanisms to prioritize different segments of the input sequence up to the current position while predicting the next token, as shown in Eq. (9).
where \(\sqrt{{d}_{k}}\) is the square root of the dimension of the embeddings, and KT is the transpose of the K vector. Since this operation is performed in a multi-head manner, the projected vectors are split into multiple heads. Therefore, the outputs of all heads are then concatenated and passed through normalization layers. Similarly, in the subsequent sublayer, Cross Multi-Head Attention is performed, which enables the decoder to focus on the generation of target-specific drugs. In this phase, QMTS is obtained from the previous MHA sublayer after normalization, and \({K}_{{{{\bf{Z}}}}_{{{\rm{Condition}}}}}\) and \({V}_{{{{\bf{Z}}}}_{{{\rm{Condition}}}}}\) are derived from the ZCondition latent vectors, where the attention is performed as shown in Eq. (10):
The resultant output is forwarded to the Residual Connection, Normalization layer, and Feed-Forward Network, through which the decoder generates the Target SMILES autoregressively as shown in Eq. (11).
where Ti is the ith generated token that belongs to the drug SMILES.
Fully-Connected Module
In this phase, the goal is to predict the affinity YAffinity between XDrugs and ZTargets by building a predictive regression model p(YAffinity∣X, ZTargets). Therefore we have concatenated the PMVO features of Graph-Encoder (Xi) and Gated-CNN module (\({Z}_{Target}^{i}\)) as Confeatures = \(({{{\bf{X}}}}^{i}| | {Z}_{Target}^{i})\). These features are then forwarded to the fully-connected layers corresponding to the affinity value Y for the affinity prediction. The affinity prediction of our proposed model can be derived as follows:
Loss functions
The loss functions of the proposed model consist of three main terms: MSE, Kullback–Leibler divergence (KL), and Language Modeling (LM) Loss. The MSE is used for the Affinity prediction task, while the KL and the LM loss are used in the generation task. The MSE calculates the mean squared difference between the actual affinities and the predicted affinity values. This operation can be expressed in Eq. (13).
where \({Y}_{i}^{\wedge }\) is the predicted affinity and Yi is the actual affinity, and n is the number of total samples.
The KL Loss aims to align the distribution of the latent space with the standard normal distribution by measuring the divergence between the approximate posterior q(z∣x) and the prior distribution p(z), which can be expressed as follows:
Here, q(z∣x) is the learned distribution by the encoders, also referred to as the approximate posterior, while p(z) is the prior distribution. More precisely, p(z) represents the assumption about the latent variables without observing any data. Since the p(z) is unobserved, therefore standard normal distribution is commonly used as p(z), such as p(z) = N(z∣0, 1), where z is the latent variable, 0 is the mean of the prior distribution, and 1 is the variance of the distribution. Moreover, d is the dimensionality of the latent space, and the \({\mu }_{i}^{2}(x)\) represents the mean of the learned distribution of each latent variable. These terms are compared during training with the mean and variance of the standard normal distribution q(z) = N(0, 1), to regularize the latent space. Further term \(\left({\sigma }_{i}^{2}(x)\right)\) prevents the model from collapsing the variance to zero.
The LM or reconstruction loss aims to ensure the correct generation of the SMILES sequence by comparing it with the target SMILES during training. This function uses cross-entropy loss to compare the predicted SMILES tokens with the target SMILES in each time step as expressed in Eq. (15).
where T is the total number of tokens in a SMILES, yt is the true label token at time step t, and p(yt∣y1:t−1, z) is the predicted probability of the token yt influenced by the previously generated token y1:t−1, while z is the latent variable from the encoder.
Further, we integrated the KL and LM loss like in Eq. (16), while the MSE is kept separate.
Further, both LMSE and LGen are provided to the FetterGrad model, where the gradient conflict between the two loss functions is checked at each step. If a conflict is detected, FetterGrad resolves it and then takes the average of the deconflicted gradients, which is passed to the Adam optimizer. If there is no conflict, the mean of both gradients is computed and provided directly to the Adam optimizer.
FetterGard model
In the DeepDTAGen, we utilized the Encoders to extract common features for affinity prediction and novel drug generation tasks in an MTL. However, MTL typically suffers from optimization issues like conflicting gradients42, where the gradients of the task objectives grow in different directions or the magnitude of one task is greater than the other task. Such problems arise when the nature of the tasks differs from one another. To tackle the gradient-conflicting issue in DTI prediction and Drug generation tasks. We developed the Fetter Gradients algorithm (FetterGard), which acts as a mediator between task gradients. In the FetterGard optimization model, we first identify whether the gradients of the two tasks are conflicting by calculating and analyzing the ED. Since the ED ranges from 0 to infinity. Therefore, we transform it into Euclidean Similarity Score (ESS), which ranges from 0 to 1 by taking advantage of inverse ED. We assume that gradients are conflicting when the ESS is less than 0.5. Secondly, we calculate the gradient Magnitude Similarity Score (MSS) (range between 0 and 1) between two gradients to identify the dominant gradient. The FetterGrad iteratively projects the dominant conflicted task gradient onto the normal plane of the other task gradient. We have characterized the occurrence of the aforementioned conditions by the following formal definitions.
Definition 1
We defined the ESSij between the gradients. We set 0.5 as the threshold value between the gradients of two tasks, gi and gj, where the gradient conflicts when the ESSij < 0.5.
Definition 2
We define the MSS between gi and gj as given by Eq. (17).
The magnitude of the gradient indicates the intensity of the rate of change between the gradients at a given point. If the absolute gradient similarity score is equal to or close to one (1) among gradients, then the gradients are similar. In other cases, the magnitude of the gradient varies exponentially if the similarity score is zero (0) or near zero.
We have two loss functions \({{{\mathcal{L}}}}_{MSE}:{{\mathbb{R}}}^{n}\to {\mathbb{R}}\) and \({{{\mathcal{L}}}}_{Gen}:{{\mathbb{R}}}^{n}\to {\mathbb{R}}\). We describe the two-task learning goal as \({{{\mathcal{L}}}}_{MSE+Gen}(\theta )={{{\mathcal{L}}}}_{MSE}(\theta )+{{{\mathcal{L}}}}_{Gen}(\theta )\) for all \(\theta \in {{\mathbb{R}}}^{n}\), where \({{{\bf{g}}}}_{1}=\nabla {{{\mathcal{L}}}}_{MSE}(\theta )\), \({{{\bf{g}}}}_{2}=\nabla {{{\mathcal{L}}}}_{Gen}(\theta )\), and g = g1 + g2. Based on these conditions, FetterGard follows the update rules reflected in Algorithm 1 for de-conflicting the gradients as the objective function.
Algorithm 1
FetterGrad Update Rule
1: Input: Model parameters θ and task mini-batch βmini = {Tn}
2: Objective: θ = θ*
3: \({g}_{n}\leftarrow {\nabla }_{\theta }{{{\mathcal{L}}}}_{n}\) for all n
4: \({g}_{n}^{\,{\mbox{FG}}\,}\leftarrow {g}_{n}\) for all n
5: for all Ti ∈ βmini do
6: for all Tj ∈ βmini do
7: if \(| {g}_{i}^{\,{\mbox{FG}}\,}-{g}_{j}| \, < \, 0.5\) then
8: Set \({g}_{i}^{{\mbox{FG}}}={g}_{i}^{{\mbox{FG}}}+| {g}_{i}-{g}_{j}| \cdot {g}_{j}\)
9: else if \(| {g}_{j}^{{\mbox{FG}}}-{g}_{1}| \, < \, 0.5\) and ∣g1∣ > gi then
10: Set \({g}_{j}^{{\mbox{FG}}}={g}_{j}^{{\mbox{FG}}}+| {g}_{j}-{g}_{i}| \cdot {g}_{i}\)
11: end if
12: end for
13: end for
14: Output: update \({{\Delta }}\theta={g}^{{{\rm{FG}}}}={\sum }_{n}{g}_{n}^{{\mbox{FG}}\,}\)
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Datasets used in this work are publicly available datasets (KIBA, Davis, and BindingDB). Source data are available with this paper as a Source Data file, and can be found along with all supporting data at: https://doi.org/10.6084/m9.figshare.28760288, including preprocessed datasets and generated drugs by DeepDTAGen. The target proteins and input drugs mentioned in Figs. 4, 5 and 8 belong to the KIBA dataset, while the mentioned target EGFR in Fig. 7 is available in the Uniprot repository (https://www.uniprot.org/) under their accession codes.
Code availability
The source code of DeepDTAGen is available on GitHub at: https://github.com/CSUBioGroup/DeepDTAGen, which is also deposited in Zenodo under accession code https://zenodo.org/records/15171802. The pre-trained models of DeepDTAGen on KIBA, Davis, and BindingDB datasets are available at https://doi.org/10.6084/m9.figshare.28760288. Alphafold 3 (https://alphafold.ebi.ac.uk/) is used to predict the 3D structures of target proteins. AutoDock VINA (https://autodock.scripps.edu/) is utilized to predict pocket area and the docking scores of the interactions. PyMol v3.1.3 (https://www.pymol.org/) is used to visualize the pocket areas for the interactions. The PyTorch v1.12.1 (https://pytorch.org/) library is used for developing the DeepDTAGen. RDKit v2022.09.1 (https://www.rdkit.org/) and ChemDraw v21.0.0 (https://revvitysignals.com/products/research/chemdraw) are utilized to obtain the chemical structure from the SMILES. Numpy v1.23.5 (https://numpy.org/) and Matplotlib v3.5.1 (https://matplotlib.org/) were used for handling and plotting the data.
References
Oprea, T. & Mestres, J. Drug repurposing: far beyond new targets for old drugs. AAPS J. 14, 759–763 (2012).
Noble, M. E., Endicott, J. A. & Johnson, L. N. Protein kinase inhibitors: insights into drug design from structure. Science 303, 1800–1805 (2004).
Lu, Z. et al. DTIAM: a unified framework for predicting drug-target interactions, binding affinities and drug mechanisms. Nat. Commun. 16, 2548 (2025).
Wang, K., Zhou, R., Li, Y. & Li, M. DeepDTAF: a deep learning method to predict protein–ligand binding affinity. Brief. Bioinform. 22, 072 (2021).
Wang, K., Zhou, R., Tang, J. & Li, M. GraphscoreDTA: optimized graph neural network for protein–ligand binding affinity prediction. Bioinformatics 39, 340 (2023).
Sun, X., Jia, X., Lu, Z., Tang, J. & Li, M. Drug repositioning with adaptive graph convolutional networks. Bioinformatics 40, 748 (2024).
Li, M., Lu, Z., Wu, Y. & Li, Y. BACPI: a bi-directional attention neural network for compound–protein interaction and binding affinity prediction. Bioinformatics 38, 1995–2002 (2022).
Bleakley, K., Biau, G. & Vert, J.-P. Supervised reconstruction of biological networks with local models. Bioinformatics 23, 57–65 (2007).
Bleakley, K. & Yamanishi, Y. Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics 25, 2397–2403 (2009).
Jacob, L. & Vert, J.-P. Protein-ligand interaction prediction: an improved chemogenomics approach. Bioinformatics 24, 2149–2156 (2008).
Mordelet, F. & Vert, J.-P. SIRENE: supervised inference of regulatory networks. Bioinformatics 24, 76–82 (2008).
Pahikkala, T. et al. Toward more realistic drug–target interaction predictions. Brief. Bioinform. 16, 325–337 (2015).
He, T., Heidemeyer, M., Ban, F., Cherkasov, A. & Ester, M. SimBoost: a read-across approach for predicting drug–target binding affinities using gradient boosting machines. J. Cheminform. 9, 1–14 (2017).
Öztürk, H., Özgür, A. & Ozkirimli, E. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics 34, 821–829 (2018).
Öztürk, H., Ozkirimli, E. & Özgür, A. Widedta: prediction of drug-target binding affinity. arXiv preprint arXiv:1902.04166 (2019).
Nguyen, T. et al. GraphDTA: predicting drug–target binding affinity with graph neural networks. Bioinformatics 37, 1140–1147 (2021).
Li, T., Zhao, X.-M. & Li, L. Co-VAE: drug-target binding affinity prediction by co-regularized variational autoencoders. IEEE Trans. Pattern Anal. Mach. Intell. 44, 8861–8873 (2021).
Abbasi, M. et al. Designing optimized drug candidates with generative adversarial network. J. Cheminform. 14, 40 (2022).
Guimaraes, G. L., Sanchez-Lengeling, B., Outeiral, C., Farias, P. L. C. & Aspuru-Guzik, A. Objective-reinforced generative adversarial networks (organ) for sequence generation models. arXiv preprint arXiv:1705.10843 (2017).
Zhu, H., Zhou, R., Cao, D., Tang, J. & Li, M. A pharmacophore-guided deep learning approach for bioactive molecular generation. Nat. Commun. 14, 6234 (2023).
Padalkar, G. R., Patil, S. D., Hegadi, M. M. & Jaybhaye, N. K. Drug discovery using generative adversarial network with reinforcement learning. In Proc. 2021 International Conference on Computer Communication and Informatics 1–3 (IEEE, 2021).
Zheng, W. et al. Federated Learning with Fair Averaging. In Proceedings of the Thirtieth International Joint Conference on Artificial Intelligence. pp. 1615–1623 (International Joint Conferences on Artificial Intelligence Organization, 2021).
Zhang, L., Zeng, W., Chen, J., Chen, J. & Li, K. GDilatedDTA: graph dilation convolution strategy for drug target binding affinity prediction. Biomed. Signal Process. Control 92, 106110 (2024).
Segler, M. H., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018).
Yang, Y. et al. Syntalinker: automatic fragment linking with deep conditional transformer neural networks. Chem. Sci. 11, 8312–8322 (2020).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46, 2699–2699 (2018).
Giménez, B., Santos, M., Ferrarini, M. & Fernandes, J. Evaluation of blockbuster drugs under the rule-of-five. Die Pharmazie Int. J. Pharm. Sci. 65, 148–152 (2010).
Normanno, N. et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366, 2–16 (2006).
Boran, A. D. & Iyengar, R. Systems pharmacology. Mt. Sinai J. Med. J. Transl. Pers. Med. 77, 333–344 (2010).
Yıldırım, M. A., Goh, K.-I., Cusick, M. E., Barabási, A.-L. & Vidal, M. Drug—target network. Nat. Biotechnol. 25, 1119–1126 (2007).
Xie, L., Xie, L., Kinnings, S. L. & Bourne, P. E. Novel computational approaches to polypharmacology as a means to define responses to individual drugs. Annu. Rev. Pharmacol. Toxicol. 52, 361–379 (2012).
Reddy, A. S. & Zhang, S. Polypharmacology: drug discovery for the future. Expert Rev. Clin. Pharmacol. 6, 41–47 (2013).
Tang, J. et al. Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J. Chem. Inf. Model. 54, 735–743 (2014).
Davis, M. I. et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1046–1051 (2011).
Liu, T., Lin, Y., Wen, X., Jorissen, R. N. & Gilson, M. K. Bindingdb: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 35, 198–201 (2007).
Bolton, E. E., Wang, Y. & Thiessen, P. A. PubChem: integrated platform of small molecules and biological activities. in Annual Reports in Computational Chemistry Vol. 4 (eds Bryant, S. H., Wheeler, R. A.) 217–241 (Elsevier, 2008).
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R. & Magrane, M. Uniprot: the universal protein knowledgebase. Nucleic Acids Res. 32, 115–119 (2004).
Landrum, G. et al. RDKit: open-source cheminformatics software (2016).
Dauphin, Y. N., Fan, A., Auli, M. & Grangier, D. Language modeling with gated convolutional networks. In Proc. International Conference on Machine Learning 933–941 (PMLR, 2017).
Kingma, D. P. & Welling, M. Stochastic gradient VB and the variational auto-encoder. In Second international conference on learning representations. Vol. 19, p. 121 (ICLR, 2014).
Yu, T., Kumar, S., Gupta, A., Levine, S., Hausman, K. & Finn, C. Gradient surgery for multi-task learning. Adv. Neural Inf. Process. Syst. 33, 5824–5836 (2020).
Zhao, Q., Duan, G., Yang, M., Cheng, Z., Li, Y. & Wang, J. AttentionDTA: drug-target binding affinity prediction by sequence-based deep learning with attention mechanism. IEEE/ACM Trans. Comput. Biol. Bioinforma. 20, 852–863 (2022).
Abbasi, K., Razzaghi, P., Poso, A., Amanlou, M., Ghasemi, J. B. & Masoudi-Nejad, A. DeepCDA: deep cross-domain compound–protein affinity prediction through LSTM and convolutional neural networks. Bioinformatics 36, 4633–4642 (2020).
Wang, J., Wen, N., Wang, C., Zhao, L. & Cheng, L. ELECTRA-DTA: a new compound-protein binding affinity prediction model based on the contextualized sequence encoding. J. Cheminform. 14, 14 (2022).
Qian, Y., Ni, W., Xianyu, X., Tao, L. & Wang, Q. DoubleSG-DTA: deep learning for drug discovery: case study on the non-small cell lung cancer with EGFR T 790 M mutation. Pharmaceutics 15, 675 (2023).
Pei, Q. et al. Breaking the barriers of data scarcity in drug–target affinity prediction. Brief. Bioinform. 24, 386 (2023).
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under Grant No. 62225209, the Project of Xiangjiang Laboratory (24XJJCYJ01003), and the Academy of Finland No. 359752. We are grateful to the High-Performance Computing Center of Central South University for partial support of this work.
Author information
Authors and Affiliations
Contributions
M.L. supervised the research. P.M.S., H.Z. and M.L. conceived the initial idea. P.M.S., K.W., Z.L. and H.Z. collected and preprocessed the data. P.M.S. designed the model and wrote the manuscript. H.Z., J.T. and M.L. aided in interpreting the results. All authors revised the manuscript and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Maha A. Thafar and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shah, P.M., Zhu, H., Lu, Z. et al. DeepDTAGen: a multitask deep learning framework for drug-target affinity prediction and target-aware drugs generation. Nat Commun 16, 5021 (2025). https://doi.org/10.1038/s41467-025-59917-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-59917-6